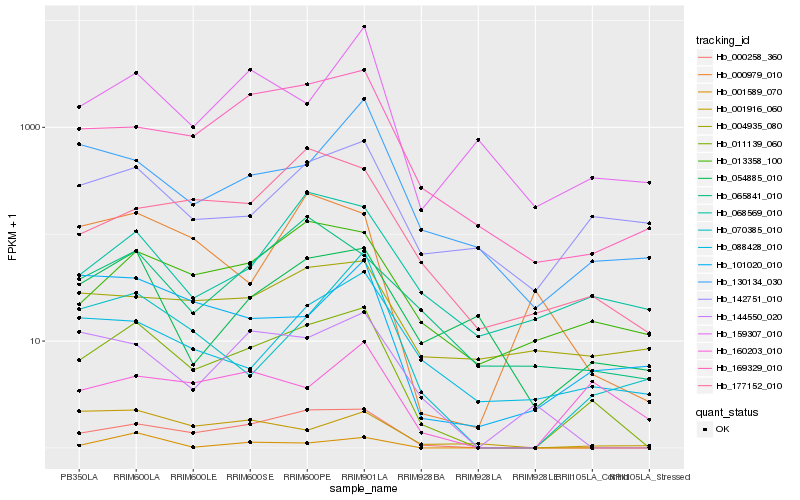
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011139_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_000258_360 |
0.1539409553 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 3 |
Hb_169329_010 |
0.2098247946 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 4 |
Hb_013358_100 |
0.2211188876 |
- |
- |
unknown [Medicago truncatula] |
| 5 |
Hb_054885_010 |
0.2383647463 |
- |
- |
- |
| 6 |
Hb_144550_020 |
0.2403974122 |
- |
- |
- |
| 7 |
Hb_177152_010 |
0.2420419412 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 8 |
Hb_160203_010 |
0.2456986329 |
- |
- |
- |
| 9 |
Hb_142751_010 |
0.2466085645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_070385_010 |
0.2481766617 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 11 |
Hb_101020_010 |
0.2486040241 |
- |
- |
- |
| 12 |
Hb_065841_010 |
0.25211211 |
- |
- |
hypothetical protein JCGZ_18376 [Jatropha curcas] |
| 13 |
Hb_001589_070 |
0.2521374985 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 14 |
Hb_159307_010 |
0.2530877457 |
- |
- |
PREDICTED: probable isoleucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 15 |
Hb_088428_010 |
0.2536583321 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 16 |
Hb_001916_060 |
0.2538732173 |
- |
- |
PREDICTED: putative protein FAR1-RELATED SEQUENCE 10 isoform X2 [Jatropha curcas] |
| 17 |
Hb_130134_030 |
0.2540108605 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 18 |
Hb_004935_080 |
0.255500973 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 19 |
Hb_000979_010 |
0.2570902259 |
- |
- |
PREDICTED: uncharacterized protein LOC105128288 [Populus euphratica] |
| 20 |
Hb_068569_010 |
0.258631882 |
- |
- |
- |