| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
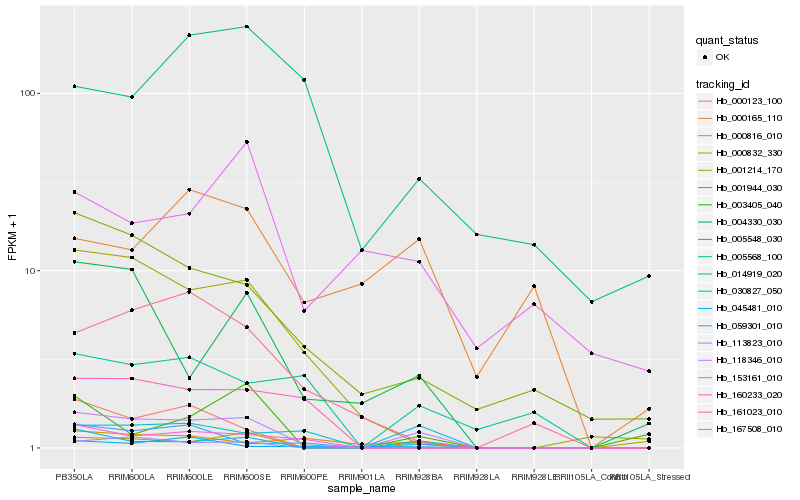
Hb_118346_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 2 |
Hb_014919_020 |
0.2430246849 |
- |
- |
- |
| 3 |
Hb_059301_010 |
0.2595042249 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 4 |
Hb_004330_030 |
0.2621038155 |
- |
- |
- |
| 5 |
Hb_001944_030 |
0.2623332668 |
- |
- |
hypothetical protein POPTR_0007s06520g [Populus trichocarpa] |
| 6 |
Hb_001214_170 |
0.2624128356 |
- |
- |
pyruvate kinase family protein [Medicago truncatula] |
| 7 |
Hb_113823_010 |
0.2874047967 |
- |
- |
- |
| 8 |
Hb_160233_020 |
0.2887696536 |
- |
- |
- |
| 9 |
Hb_167508_010 |
0.2976251074 |
- |
- |
hypothetical protein POPTR_0001s27330g [Populus trichocarpa] |
| 10 |
Hb_000832_330 |
0.2981758551 |
- |
- |
PREDICTED: uncharacterized protein LOC102661627 [Glycine max] |
| 11 |
Hb_000123_100 |
0.2988177201 |
- |
- |
Uncharacterized protein TCM_024608 [Theobroma cacao] |
| 12 |
Hb_005568_100 |
0.3040582334 |
- |
- |
vacuolar proton ATPase a3-like protein [Medicago truncatula] |
| 13 |
Hb_000816_010 |
0.3044715548 |
- |
- |
PREDICTED: uncharacterized protein At2g40430 isoform X2 [Jatropha curcas] |
| 14 |
Hb_161023_010 |
0.3074530924 |
- |
- |
PREDICTED: uncharacterized protein LOC105639929 [Jatropha curcas] |
| 15 |
Hb_030827_050 |
0.3129182485 |
- |
- |
hypothetical protein CISIN_1g040641mg, partial [Citrus sinensis] |
| 16 |
Hb_003405_040 |
0.3154406353 |
- |
- |
PREDICTED: uncharacterized protein LOC105121978 [Populus euphratica] |
| 17 |
Hb_045481_010 |
0.3237248296 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_005548_030 |
0.3355557469 |
- |
- |
hypothetical protein JCGZ_05606 [Jatropha curcas] |
| 19 |
Hb_000165_110 |
0.3360059598 |
- |
- |
unknown [Medicago truncatula] |
| 20 |
Hb_153161_010 |
0.3360827031 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |