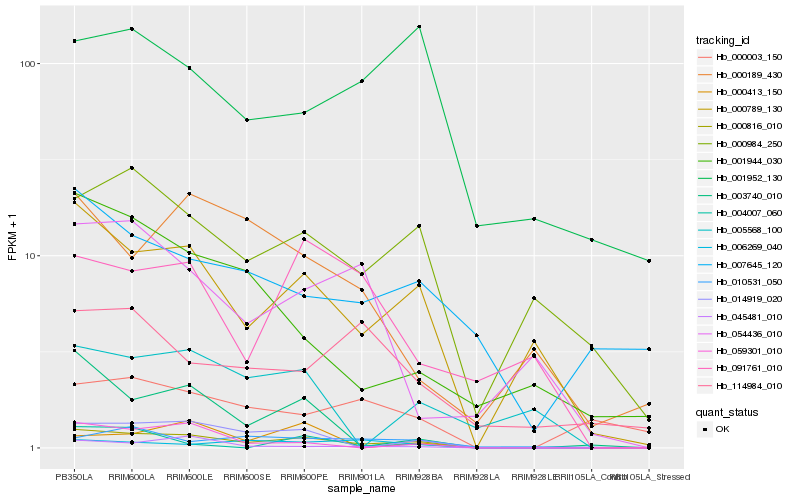
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At2g40430 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000789_130 |
0.1696155882 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 3 |
Hb_014919_020 |
0.1947809622 |
- |
- |
- |
| 4 |
Hb_000984_250 |
0.2303976722 |
- |
- |
PREDICTED: uncharacterized protein LOC105630783 [Jatropha curcas] |
| 5 |
Hb_003740_010 |
0.2328919712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_059301_010 |
0.2414245 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 7 |
Hb_045481_010 |
0.2436949008 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_007645_120 |
0.2651650771 |
- |
- |
hypothetical protein POPTR_0006s05370g [Populus trichocarpa] |
| 9 |
Hb_001944_030 |
0.2667565847 |
- |
- |
hypothetical protein POPTR_0007s06520g [Populus trichocarpa] |
| 10 |
Hb_006269_040 |
0.2678341569 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 11 |
Hb_005568_100 |
0.2688539221 |
- |
- |
vacuolar proton ATPase a3-like protein [Medicago truncatula] |
| 12 |
Hb_000189_430 |
0.2729772977 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_091761_010 |
0.2757263664 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 14 |
Hb_001952_130 |
0.2760337734 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 15 |
Hb_054436_010 |
0.2762491469 |
- |
- |
PREDICTED: uncharacterized protein LOC105638666 [Jatropha curcas] |
| 16 |
Hb_000003_150 |
0.276463208 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 6 [Jatropha curcas] |
| 17 |
Hb_010531_050 |
0.2789838794 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 18 |
Hb_114984_010 |
0.2848440601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000413_150 |
0.2850139284 |
- |
- |
PREDICTED: uncharacterized protein LOC103709996 [Phoenix dactylifera] |
| 20 |
Hb_004007_060 |
0.2869830255 |
- |
- |
Ribose-5-phosphate isomerase A [Gossypium arboreum] |