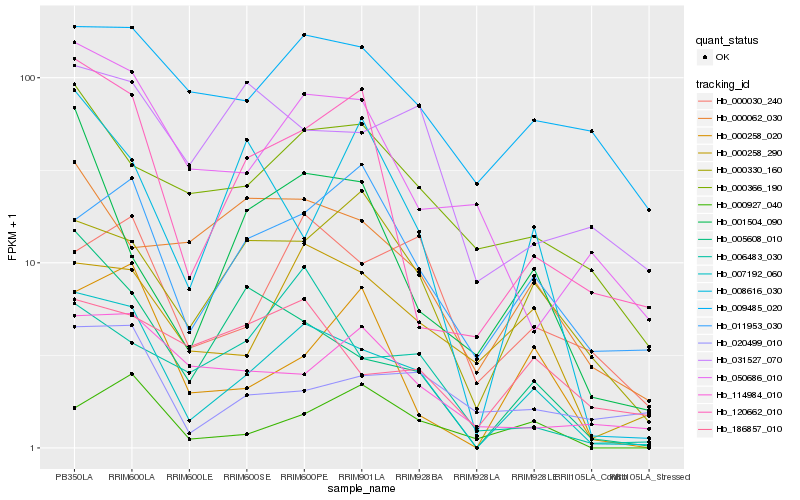
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007192_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005608_010 |
0.2140075604 |
- |
- |
PREDICTED: U-box domain-containing protein 40 [Jatropha curcas] |
| 3 |
Hb_000258_290 |
0.2186818712 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP15 [Jatropha curcas] |
| 4 |
Hb_120662_010 |
0.2242941309 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 5 |
Hb_000030_240 |
0.2346631154 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_050686_010 |
0.2356564812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000366_190 |
0.2414321879 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 8 |
Hb_000258_020 |
0.2440502099 |
- |
- |
PREDICTED: RING-H2 finger protein ATL74 [Jatropha curcas] |
| 9 |
Hb_000330_160 |
0.2446436385 |
- |
- |
hypothetical protein JCGZ_04462 [Jatropha curcas] |
| 10 |
Hb_009485_020 |
0.2456516986 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 11 |
Hb_020499_010 |
0.2512562799 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_186857_010 |
0.2517587183 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 13 |
Hb_011953_030 |
0.2521191372 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0003s14450g [Populus trichocarpa] |
| 14 |
Hb_000062_030 |
0.2562010595 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 15 |
Hb_114984_010 |
0.2566917865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001504_090 |
0.2581211079 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_000927_040 |
0.2586173748 |
- |
- |
- |
| 18 |
Hb_006483_030 |
0.2594930937 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 19 |
Hb_031527_070 |
0.2595145988 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 20 |
Hb_008616_030 |
0.2612098236 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |