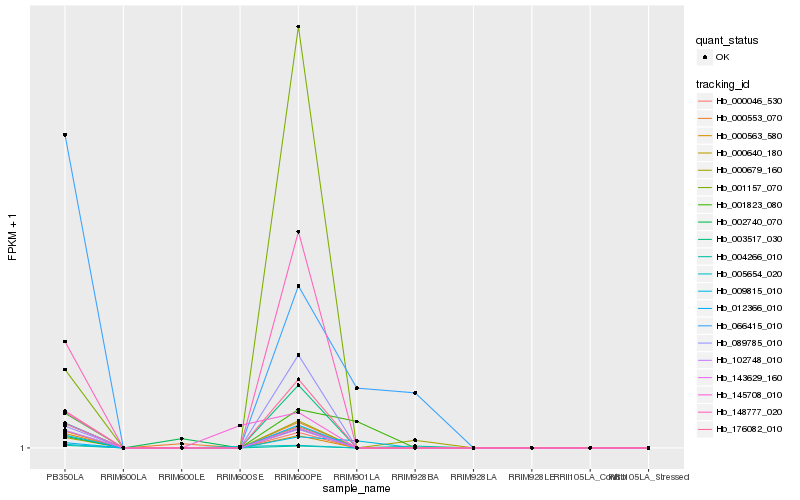
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_580 |
0.0 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000640_180 |
0.0070884437 |
- |
- |
enzyme inhibitor, putative [Ricinus communis] |
| 3 |
Hb_000046_530 |
0.0151633787 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF086 [Jatropha curcas] |
| 4 |
Hb_102748_010 |
0.0465854149 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 5 |
Hb_143629_160 |
0.0880568838 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 6 |
Hb_176082_010 |
0.1706301492 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 7 |
Hb_148777_020 |
0.207898731 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 isoform X1 [Populus euphratica] |
| 8 |
Hb_003517_030 |
0.2309828702 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 5 isoform X2 [Eucalyptus grandis] |
| 9 |
Hb_000553_070 |
0.2467818378 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X2 [Jatropha curcas] |
| 10 |
Hb_066415_010 |
0.3188428452 |
- |
- |
- |
| 11 |
Hb_012366_010 |
0.3195183279 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 12 |
Hb_089785_010 |
0.3273078834 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Fragaria vesca subsp. vesca] |
| 13 |
Hb_005654_020 |
0.3301768651 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 14 |
Hb_009815_010 |
0.3523403938 |
- |
- |
- |
| 15 |
Hb_000679_160 |
0.3701642589 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 16 |
Hb_002740_070 |
0.3781477733 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 17 |
Hb_001823_080 |
0.3862449146 |
- |
- |
hypothetical protein VITISV_012142 [Vitis vinifera] |
| 18 |
Hb_145708_010 |
0.4022750703 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_004266_010 |
0.4031649873 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 20 |
Hb_001157_070 |
0.4062401422 |
- |
- |
- |