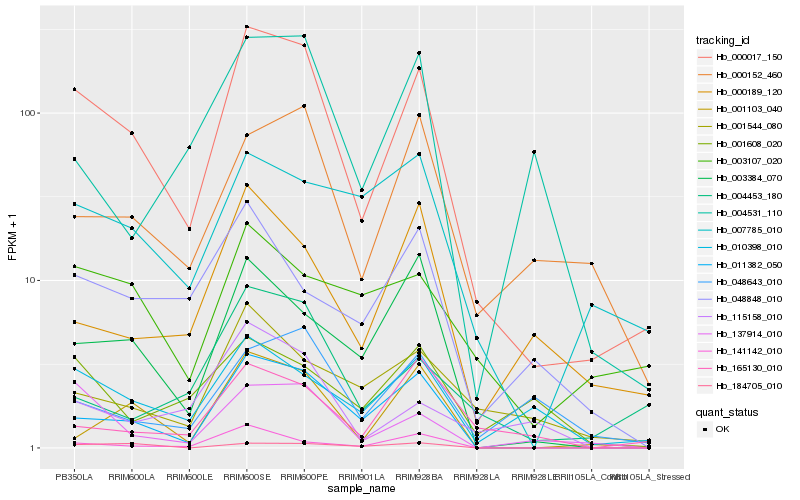
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000017_150 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s09780g [Populus trichocarpa] |
| 2 |
Hb_011382_050 |
0.1665441762 |
- |
- |
hypothetical protein PRUPE_ppa025935mg [Prunus persica] |
| 3 |
Hb_010398_010 |
0.2093350021 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 4 |
Hb_137914_010 |
0.2131925985 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 5 |
Hb_003384_070 |
0.2175923976 |
- |
- |
PREDICTED: methylesterase 17-like [Populus euphratica] |
| 6 |
Hb_000152_460 |
0.2279163429 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 7 |
Hb_001103_040 |
0.230785673 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 8 |
Hb_141142_010 |
0.2323027525 |
- |
- |
hypothetical protein JCGZ_21320 [Jatropha curcas] |
| 9 |
Hb_184705_010 |
0.2412492458 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007785_010 |
0.2413676775 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 11 |
Hb_001544_080 |
0.2415231421 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 12 |
Hb_000189_120 |
0.2434881572 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 13 |
Hb_003107_020 |
0.2435925874 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 14 |
Hb_004453_180 |
0.2443253331 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 15 |
Hb_115158_010 |
0.2469872997 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 16 |
Hb_048643_010 |
0.2485301638 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_048848_010 |
0.2489635016 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 18 |
Hb_165130_010 |
0.2526706375 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 19 |
Hb_004531_110 |
0.2557627481 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 20 |
Hb_001608_020 |
0.2582558465 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |