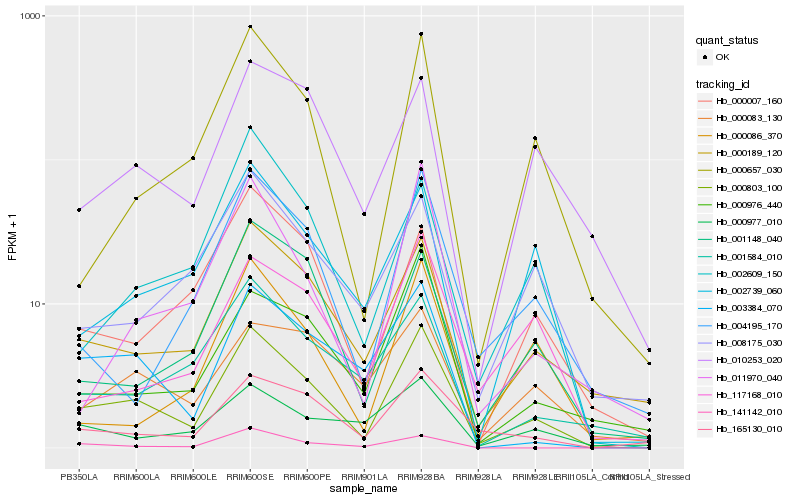
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 2 |
Hb_000189_120 |
0.1566234075 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 3 |
Hb_000657_030 |
0.1834351238 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 4 |
Hb_003384_070 |
0.1854587958 |
- |
- |
PREDICTED: methylesterase 17-like [Populus euphratica] |
| 5 |
Hb_165130_010 |
0.1856462795 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 6 |
Hb_002739_060 |
0.1931045663 |
- |
- |
- |
| 7 |
Hb_010253_020 |
0.1976758501 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_004195_170 |
0.197713803 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_001148_040 |
0.1984795978 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 10 |
Hb_000976_440 |
0.1987673838 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000007_160 |
0.1995445687 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 12 |
Hb_011970_040 |
0.2016942898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001584_010 |
0.2018033547 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 14 |
Hb_141142_010 |
0.2033769477 |
- |
- |
hypothetical protein JCGZ_21320 [Jatropha curcas] |
| 15 |
Hb_002609_150 |
0.204628818 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_117168_010 |
0.2055206805 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 17 |
Hb_000083_130 |
0.2072964085 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 18 |
Hb_000086_370 |
0.2084213468 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 19 |
Hb_008175_030 |
0.2084840008 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_000977_010 |
0.210137194 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |