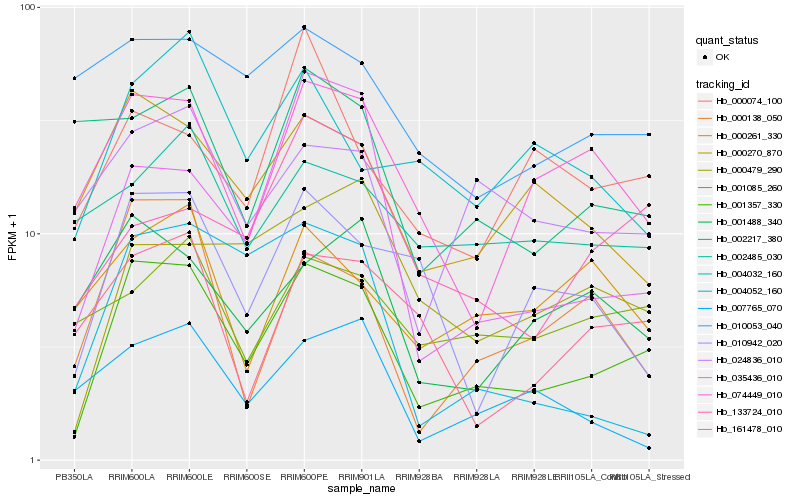
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_330 |
0.0 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 2 |
Hb_000270_870 |
0.1898857076 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 3 |
Hb_074449_010 |
0.1934936402 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 4 |
Hb_024836_010 |
0.2011873297 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 5 |
Hb_000138_050 |
0.2030713158 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper protein HAT14, putative [Ricinus communis] |
| 6 |
Hb_010942_020 |
0.2050868493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_161478_010 |
0.2090730301 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000479_290 |
0.2115406804 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001488_340 |
0.213232893 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 10 |
Hb_004032_160 |
0.2184154225 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 11 |
Hb_001085_260 |
0.2186227491 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 12 |
Hb_035436_010 |
0.2192712365 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 13 |
Hb_002485_030 |
0.2216114135 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 14 |
Hb_010053_040 |
0.2257841224 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 15 |
Hb_000261_330 |
0.2262705387 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Amborella trichopoda] |
| 16 |
Hb_133724_010 |
0.2269656024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004052_160 |
0.2278829978 |
- |
- |
Chain A, Rna Binding Protein |
| 18 |
Hb_002217_380 |
0.2290286266 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 19 |
Hb_007765_070 |
0.2290626674 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |
| 20 |
Hb_000074_100 |
0.2314844854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |