| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
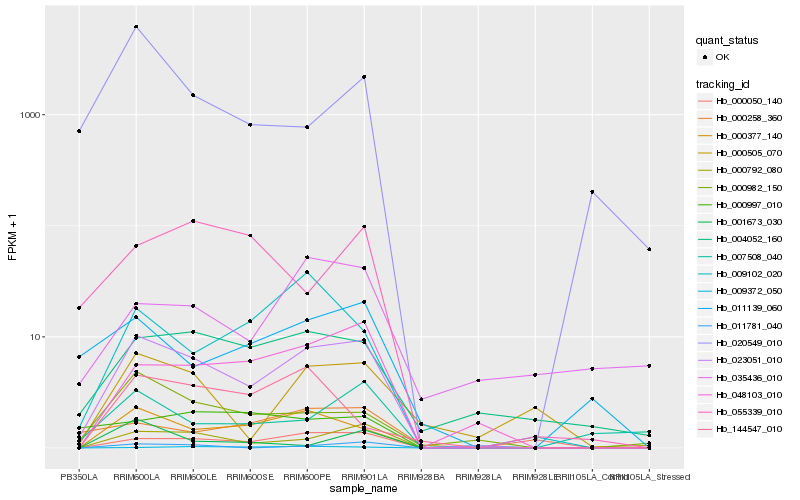
Hb_023051_010 |
0.0 |
- |
- |
PREDICTED: putative amidase C869.01 [Populus euphratica] |
| 2 |
Hb_000792_080 |
0.1689383883 |
- |
- |
hypothetical protein B456_012G111300, partial [Gossypium raimondii] |
| 3 |
Hb_000377_140 |
0.199467457 |
- |
- |
- |
| 4 |
Hb_048103_010 |
0.2158598284 |
- |
- |
- |
| 5 |
Hb_004052_160 |
0.2367694854 |
- |
- |
Chain A, Rna Binding Protein |
| 6 |
Hb_000982_150 |
0.2381217844 |
transcription factor |
TF Family: Orphans |
histidine kinase 1 plant, putative [Ricinus communis] |
| 7 |
Hb_011781_040 |
0.2529520979 |
- |
- |
- |
| 8 |
Hb_000258_360 |
0.2610648824 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 9 |
Hb_011139_060 |
0.2641319159 |
- |
- |
- |
| 10 |
Hb_000505_070 |
0.2727841654 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009102_020 |
0.2758812931 |
- |
- |
hypothetical protein EUGRSUZ_I01262 [Eucalyptus grandis] |
| 12 |
Hb_009372_050 |
0.2820067569 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_007508_040 |
0.2870675381 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000997_010 |
0.289413544 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000050_140 |
0.2917366442 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 isoform X1 [Populus euphratica] |
| 16 |
Hb_055339_010 |
0.2956099089 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 17 |
Hb_020549_010 |
0.2973302153 |
- |
- |
PREDICTED: uncharacterized protein LOC105638664 isoform X2 [Jatropha curcas] |
| 18 |
Hb_035436_010 |
0.3009901156 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 19 |
Hb_144547_010 |
0.3034849614 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 20 |
Hb_001673_030 |
0.3048222962 |
- |
- |
hypothetical protein JCGZ_08729 [Jatropha curcas] |