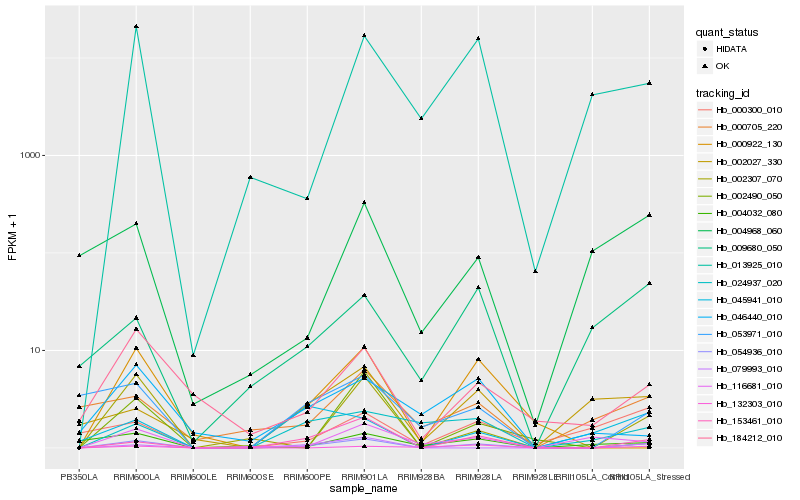
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054936_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 2 |
Hb_153461_010 |
0.2552405436 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 3 |
Hb_002307_070 |
0.2907588474 |
- |
- |
hypothetical protein JCGZ_14841 [Jatropha curcas] |
| 4 |
Hb_184212_010 |
0.3193596623 |
- |
- |
PREDICTED: uncharacterized protein LOC105113658 [Populus euphratica] |
| 5 |
Hb_013925_010 |
0.3215366018 |
- |
- |
RecName: Full=Pro-hevein; AltName: Full=Major hevein; Contains: RecName: Full=Hevein; AltName: Allergen=Hev b 6; Contains: RecName: Full=Win-like protein; Flags: Precursor [Hevea brasiliensis] |
| 6 |
Hb_079993_010 |
0.3330976353 |
- |
- |
PREDICTED: uncharacterized protein LOC103336399 [Prunus mume] |
| 7 |
Hb_000922_130 |
0.3357115017 |
- |
- |
hypothetical protein POPTR_0008s04925g [Populus trichocarpa] |
| 8 |
Hb_053971_010 |
0.3382606177 |
- |
- |
- |
| 9 |
Hb_116681_010 |
0.3508349585 |
- |
- |
hypothetical protein PRUPE_ppa025968mg, partial [Prunus persica] |
| 10 |
Hb_004032_080 |
0.3552896813 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL11 [Jatropha curcas] |
| 11 |
Hb_024937_020 |
0.3566176336 |
- |
- |
- |
| 12 |
Hb_132303_010 |
0.3588648688 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_002027_330 |
0.3640614002 |
- |
- |
- |
| 14 |
Hb_002490_050 |
0.365035567 |
- |
- |
Os02g0182500, partial [Oryza sativa Japonica Group] |
| 15 |
Hb_046440_010 |
0.3663759207 |
- |
- |
hypothetical protein JCGZ_06190 [Jatropha curcas] |
| 16 |
Hb_004968_060 |
0.3702243973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000705_220 |
0.3709792392 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_009680_050 |
0.3715579799 |
- |
- |
- |
| 19 |
Hb_045941_010 |
0.3745022395 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 20 |
Hb_000300_010 |
0.3775921006 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |