| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
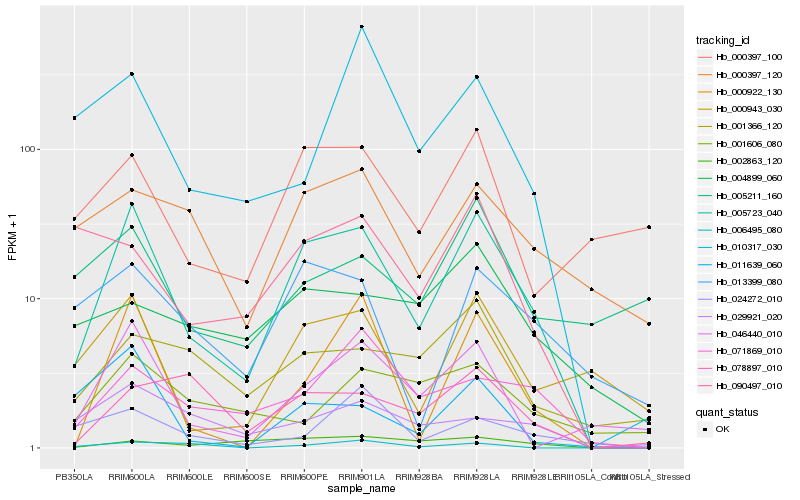
Hb_005723_040 |
0.0 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_046440_010 |
0.2363292995 |
- |
- |
hypothetical protein JCGZ_06190 [Jatropha curcas] |
| 3 |
Hb_000943_030 |
0.2388676796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000922_130 |
0.2432333353 |
- |
- |
hypothetical protein POPTR_0008s04925g [Populus trichocarpa] |
| 5 |
Hb_013399_080 |
0.2660022885 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 6 |
Hb_071869_010 |
0.2676900345 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000397_100 |
0.2736274744 |
- |
- |
hypothetical protein 31 [Hevea brasiliensis] |
| 8 |
Hb_000397_120 |
0.2778370771 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 9 |
Hb_001366_120 |
0.2793653132 |
- |
- |
unknown [Zea mays] |
| 10 |
Hb_011639_060 |
0.2800946632 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 11 |
Hb_078897_010 |
0.2830689849 |
- |
- |
hypothetical protein POPTR_0012s044902g, partial [Populus trichocarpa] |
| 12 |
Hb_090497_010 |
0.2856362368 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 13 |
Hb_029921_020 |
0.2862166968 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 14 |
Hb_002863_120 |
0.2885800581 |
desease resistance |
Gene Name: AAA_17 |
JHL25H03.3 [Jatropha curcas] |
| 15 |
Hb_010317_030 |
0.297813327 |
- |
- |
unknown [Picea sitchensis] |
| 16 |
Hb_006495_080 |
0.2982473259 |
- |
- |
hypothetical protein PRUPE_ppb025438mg, partial [Prunus persica] |
| 17 |
Hb_024272_010 |
0.3004110687 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 18 |
Hb_004899_060 |
0.3010851885 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 19 |
Hb_005211_160 |
0.3012654671 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 20 |
Hb_001606_080 |
0.3025359186 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X2 [Jatropha curcas] |