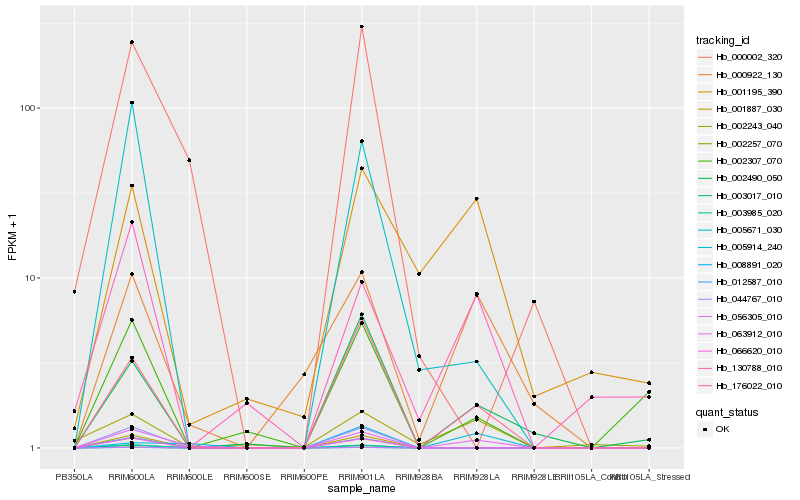
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063912_010 |
0.0 |
- |
- |
PREDICTED: elongation factor 1-alpha-like, partial [Cucumis sativus] |
| 2 |
Hb_176022_010 |
0.2380170504 |
- |
- |
- |
| 3 |
Hb_000922_130 |
0.2471346024 |
- |
- |
hypothetical protein POPTR_0008s04925g [Populus trichocarpa] |
| 4 |
Hb_130788_010 |
0.2947079492 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 5 |
Hb_002490_050 |
0.2951624257 |
- |
- |
Os02g0182500, partial [Oryza sativa Japonica Group] |
| 6 |
Hb_001195_390 |
0.305573763 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Populus euphratica] |
| 7 |
Hb_005671_030 |
0.3087352759 |
- |
- |
- |
| 8 |
Hb_002243_040 |
0.3090777812 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00227 [Jatropha curcas] |
| 9 |
Hb_005914_240 |
0.326149926 |
- |
- |
PREDICTED: oxidation resistance protein 1 [Jatropha curcas] |
| 10 |
Hb_000002_320 |
0.330568949 |
- |
- |
PREDICTED: uncharacterized protein LOC105632108 [Jatropha curcas] |
| 11 |
Hb_002307_070 |
0.3331615946 |
- |
- |
hypothetical protein JCGZ_14841 [Jatropha curcas] |
| 12 |
Hb_008891_020 |
0.3371770928 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 13 |
Hb_056305_010 |
0.3371785351 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_002257_070 |
0.3371848768 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 15 |
Hb_003017_010 |
0.3372047232 |
- |
- |
PREDICTED: uncharacterized protein LOC104878882 [Vitis vinifera] |
| 16 |
Hb_003985_020 |
0.3372728677 |
- |
- |
- |
| 17 |
Hb_012587_010 |
0.3374065731 |
- |
- |
- |
| 18 |
Hb_066620_010 |
0.3377837546 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |
| 19 |
Hb_044767_010 |
0.3379644201 |
- |
- |
- |
| 20 |
Hb_001887_030 |
0.338178484 |
- |
- |
- |