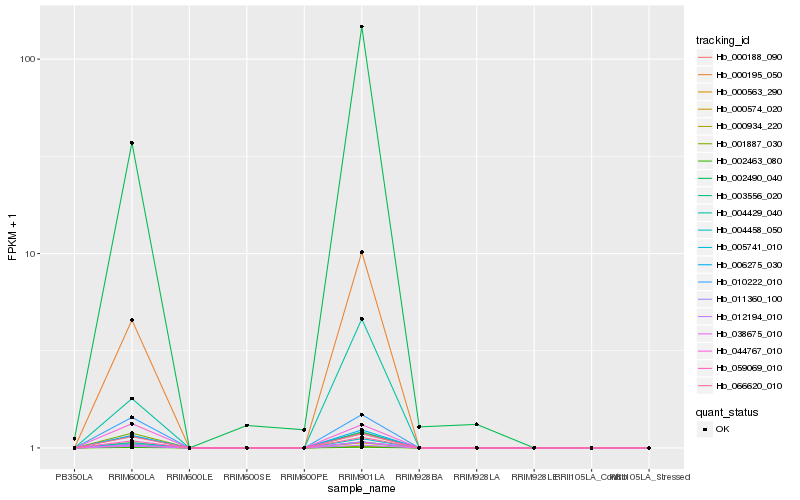
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_059069_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105180180, partial [Sesamum indicum] |
| 2 |
Hb_038675_010 |
0.0011207789 |
- |
- |
PREDICTED: uncharacterized protein LOC105155307, partial [Sesamum indicum] |
| 3 |
Hb_011360_100 |
0.0167076207 |
- |
- |
- |
| 4 |
Hb_004458_050 |
0.0306946525 |
- |
- |
- |
| 5 |
Hb_005741_010 |
0.0354384449 |
- |
- |
PREDICTED: uncharacterized protein LOC105775353 [Gossypium raimondii] |
| 6 |
Hb_000195_050 |
0.0401032941 |
- |
- |
- |
| 7 |
Hb_012194_010 |
0.0480269196 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 8 |
Hb_006275_030 |
0.0701848356 |
- |
- |
- |
| 9 |
Hb_000574_020 |
0.0732442204 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000563_290 |
0.0742782375 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 11 |
Hb_002463_080 |
0.0809908672 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_003556_020 |
0.0842252351 |
- |
- |
PREDICTED: non-specific lipid-transfer protein D, cotyledon-specific isoform-like [Citrus sinensis] |
| 13 |
Hb_010222_010 |
0.1316327381 |
- |
- |
- |
| 14 |
Hb_002490_040 |
0.1339865139 |
- |
- |
20S proteasome beta subunit C2 [Hevea brasiliensis] |
| 15 |
Hb_000188_090 |
0.1348972268 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 16 |
Hb_004429_040 |
0.1399999471 |
- |
- |
- |
| 17 |
Hb_000934_220 |
0.140600434 |
- |
- |
PREDICTED: uncharacterized protein LOC104890722 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_001887_030 |
0.1603914538 |
- |
- |
- |
| 19 |
Hb_044767_010 |
0.1634006467 |
- |
- |
- |
| 20 |
Hb_066620_010 |
0.1662499717 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |