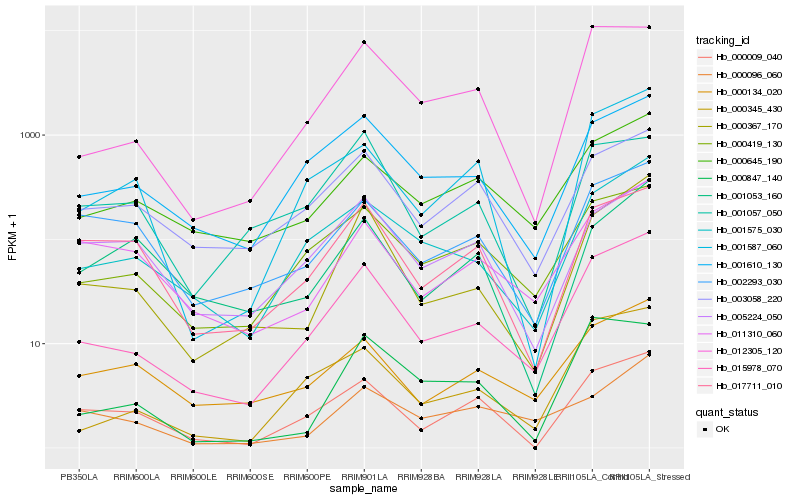
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_170 |
0.0 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |
| 2 |
Hb_015978_070 |
0.1152560483 |
- |
- |
- |
| 3 |
Hb_001575_030 |
0.1689115529 |
- |
- |
hypothetical protein POPTR_0022s00880g [Populus trichocarpa] |
| 4 |
Hb_001610_130 |
0.1791994687 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 5 |
Hb_011310_060 |
0.1861129292 |
- |
- |
PREDICTED: uncharacterized protein At2g34160-like [Jatropha curcas] |
| 6 |
Hb_001053_160 |
0.1882230228 |
- |
- |
hypothetical protein POPTR_0009s06660g [Populus trichocarpa] |
| 7 |
Hb_000009_040 |
0.1885256956 |
- |
- |
- |
| 8 |
Hb_000134_020 |
0.1896319661 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 9 |
Hb_001057_050 |
0.193613677 |
- |
- |
hypothetical protein POPTR_0003s15960g, partial [Populus trichocarpa] |
| 10 |
Hb_017711_010 |
0.1937550462 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM9-2 [Jatropha curcas] |
| 11 |
Hb_005224_050 |
0.1949601909 |
- |
- |
PREDICTED: uncharacterized protein LOC105632350 [Jatropha curcas] |
| 12 |
Hb_003058_220 |
0.1967348134 |
- |
- |
Pre-mRNA branch site protein p14, putative [Ricinus communis] |
| 13 |
Hb_000645_190 |
0.1982391708 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 14 |
Hb_012305_120 |
0.1985420704 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 15 |
Hb_002293_030 |
0.2002654176 |
- |
- |
eukaryotic translation initiation factor 5A isoform VI [Hevea brasiliensis] |
| 16 |
Hb_001587_060 |
0.2021053835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000096_060 |
0.2038964745 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000847_140 |
0.2094001063 |
- |
- |
- |
| 19 |
Hb_000345_430 |
0.2101753017 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 20 |
Hb_000419_130 |
0.2113255984 |
- |
- |
PREDICTED: uncharacterized protein LOC105650901 [Jatropha curcas] |