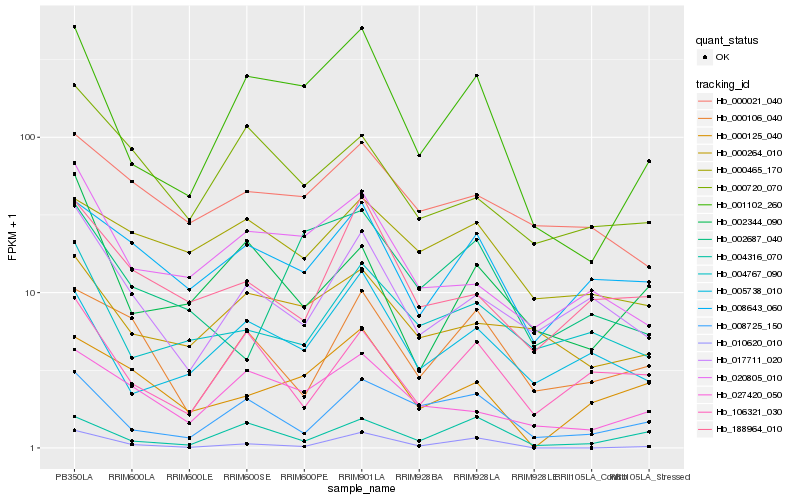
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_260 |
0.0 |
- |
- |
hypothetical protein L484_019841 [Morus notabilis] |
| 2 |
Hb_005738_010 |
0.180615087 |
- |
- |
- |
| 3 |
Hb_027420_050 |
0.191339776 |
- |
- |
- |
| 4 |
Hb_020805_010 |
0.1919863702 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_008725_150 |
0.1920052079 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 6 |
Hb_000264_010 |
0.1963007364 |
- |
- |
PREDICTED: uncharacterized protein LOC105649505 [Jatropha curcas] |
| 7 |
Hb_004316_070 |
0.200024854 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 8 |
Hb_008643_060 |
0.2168076595 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_010620_010 |
0.2184147431 |
- |
- |
hypothetical protein JCGZ_22570 [Jatropha curcas] |
| 10 |
Hb_004767_090 |
0.2267859707 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 11 |
Hb_017711_020 |
0.2277121412 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002344_090 |
0.2284351238 |
- |
- |
PREDICTED: uncharacterized protein LOC105637976 [Jatropha curcas] |
| 13 |
Hb_106321_030 |
0.2317915598 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 14 |
Hb_002687_040 |
0.2354379279 |
- |
- |
PREDICTED: alpha-galactosidase 1-like [Jatropha curcas] |
| 15 |
Hb_000720_070 |
0.2356228696 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 16 |
Hb_000125_040 |
0.2359127743 |
- |
- |
- |
| 17 |
Hb_000465_170 |
0.2363426401 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 18 |
Hb_000021_040 |
0.2367523859 |
- |
- |
PREDICTED: villin-3-like isoform X1 [Populus euphratica] |
| 19 |
Hb_188964_010 |
0.2387658959 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 20 |
Hb_000106_040 |
0.2399860987 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Vitis vinifera] |