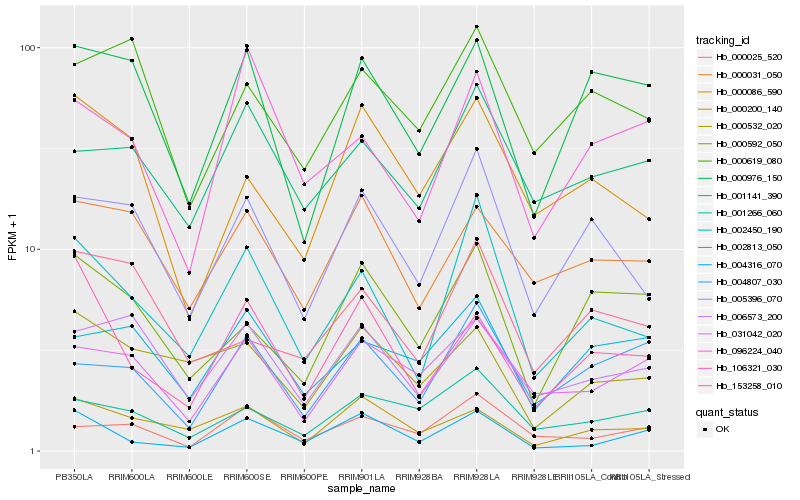
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002450_190 |
0.0 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 2 |
Hb_005396_070 |
0.1290092258 |
- |
- |
hypothetical protein EUGRSUZ_B03067 [Eucalyptus grandis] |
| 3 |
Hb_001141_390 |
0.1726556933 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 4 |
Hb_000025_520 |
0.1825796082 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27110 [Jatropha curcas] |
| 5 |
Hb_096224_040 |
0.1841966497 |
- |
- |
PREDICTED: probable nucleoredoxin 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006573_200 |
0.1872112525 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g52850, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004807_030 |
0.1892886187 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 8 |
Hb_000976_150 |
0.1909624983 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45 [Jatropha curcas] |
| 9 |
Hb_153258_010 |
0.1929339292 |
- |
- |
PREDICTED: uncharacterized protein LOC105642325 [Jatropha curcas] |
| 10 |
Hb_001266_060 |
0.1929483436 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g47840 [Jatropha curcas] |
| 11 |
Hb_004316_070 |
0.1929926133 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 12 |
Hb_031042_020 |
0.1934946372 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g47460 [Jatropha curcas] |
| 13 |
Hb_000532_020 |
0.1938785441 |
- |
- |
beta-tubulin [Bouteloua eriopoda] |
| 14 |
Hb_000031_050 |
0.1942103998 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 15 |
Hb_002813_050 |
0.1972616774 |
desease resistance |
Gene Name: TIR_2 |
PREDICTED: uncharacterized protein LOC105635699 [Jatropha curcas] |
| 16 |
Hb_000200_140 |
0.1986322907 |
- |
- |
PREDICTED: uncharacterized protein LOC104591989 [Nelumbo nucifera] |
| 17 |
Hb_000592_050 |
0.1991958438 |
- |
- |
PREDICTED: golgin candidate 2 [Jatropha curcas] |
| 18 |
Hb_000086_590 |
0.1999685593 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 19 |
Hb_106321_030 |
0.2001234925 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 20 |
Hb_000619_080 |
0.2003505746 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 21 [Jatropha curcas] |