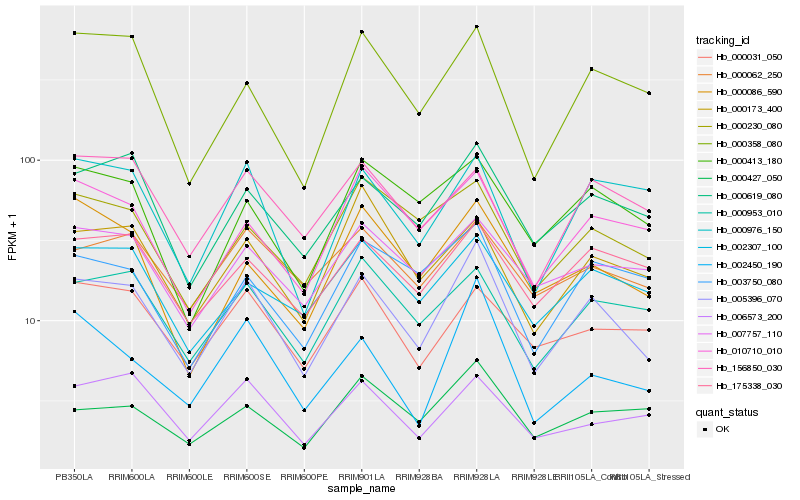
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005396_070 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_B03067 [Eucalyptus grandis] |
| 2 |
Hb_000619_080 |
0.1125263055 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 21 [Jatropha curcas] |
| 3 |
Hb_000953_010 |
0.1267750112 |
- |
- |
hypothetical protein CISIN_1g0425012mg, partial [Citrus sinensis] |
| 4 |
Hb_000358_080 |
0.1281475389 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 5 |
Hb_175338_030 |
0.129008979 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 6 |
Hb_002450_190 |
0.1290092258 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 7 |
Hb_002307_100 |
0.1297190764 |
- |
- |
PREDICTED: protein transport protein sec23-1 [Jatropha curcas] |
| 8 |
Hb_000062_250 |
0.1325089769 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 9 |
Hb_000976_150 |
0.1328645743 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45 [Jatropha curcas] |
| 10 |
Hb_000230_080 |
0.1329360783 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_007757_110 |
0.132977188 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 12 |
Hb_006573_200 |
0.1335388648 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g52850, chloroplastic [Jatropha curcas] |
| 13 |
Hb_010710_010 |
0.1336761387 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_000413_180 |
0.1361057568 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 15 |
Hb_000031_050 |
0.137992613 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 16 |
Hb_000427_050 |
0.1380372377 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_156850_030 |
0.1390368085 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 18 |
Hb_000086_590 |
0.1390691092 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003750_080 |
0.1393096899 |
- |
- |
PREDICTED: DNA repair protein complementing XP-C cells homolog [Jatropha curcas] |
| 20 |
Hb_000173_400 |
0.1413537767 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18950 [Jatropha curcas] |