| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
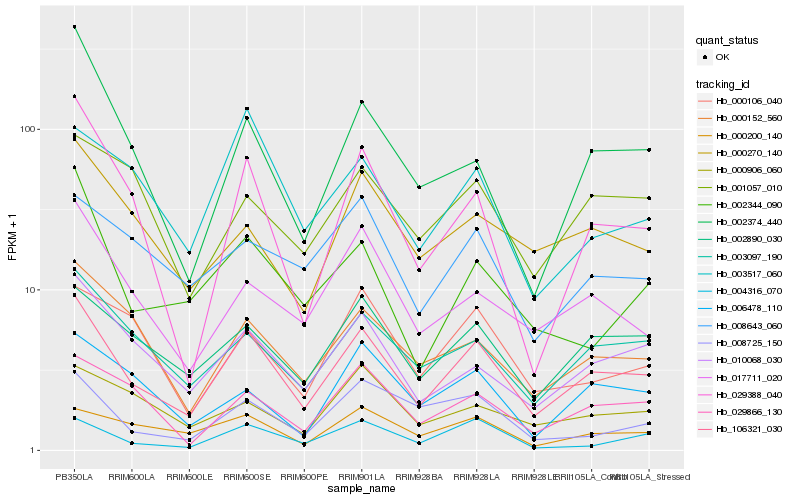
Hb_106321_030 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 2 |
Hb_002890_030 |
0.1276941456 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 3 |
Hb_003097_190 |
0.1309845372 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 4 |
Hb_010068_030 |
0.1365860632 |
- |
- |
aconitase, putative [Ricinus communis] |
| 5 |
Hb_000152_560 |
0.1417068176 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 6 |
Hb_029866_130 |
0.1447064036 |
- |
- |
hypothetical protein JCGZ_00035 [Jatropha curcas] |
| 7 |
Hb_029388_040 |
0.1492933719 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 8 |
Hb_017711_020 |
0.1500901809 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000270_140 |
0.15293948 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000200_140 |
0.1547690186 |
- |
- |
PREDICTED: uncharacterized protein LOC104591989 [Nelumbo nucifera] |
| 11 |
Hb_004316_070 |
0.1568892134 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 12 |
Hb_006478_110 |
0.1603332453 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 13 |
Hb_008643_060 |
0.1628743574 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000106_040 |
0.1644742957 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Vitis vinifera] |
| 15 |
Hb_001057_010 |
0.1645400937 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 16 |
Hb_008725_150 |
0.1671202485 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 17 |
Hb_002344_090 |
0.1674421851 |
- |
- |
PREDICTED: uncharacterized protein LOC105637976 [Jatropha curcas] |
| 18 |
Hb_002374_440 |
0.1678470545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000906_060 |
0.1688141405 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 20 |
Hb_003517_060 |
0.1711238744 |
- |
- |
hypothetical protein CISIN_1g044999mg, partial [Citrus sinensis] |