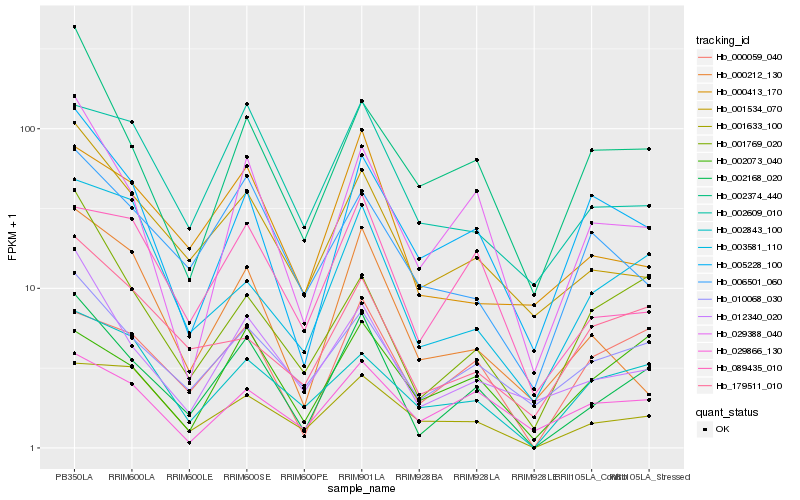
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002168_020 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_029388_040 |
0.1577127678 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 3 |
Hb_012340_020 |
0.1709439518 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 4 |
Hb_010068_030 |
0.1735005351 |
- |
- |
aconitase, putative [Ricinus communis] |
| 5 |
Hb_001534_070 |
0.1789489303 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 6 |
Hb_000059_040 |
0.1816339518 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 7 |
Hb_005228_100 |
0.1866746422 |
- |
- |
- |
| 8 |
Hb_002374_440 |
0.192713825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_089435_010 |
0.1932954543 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Vitis vinifera] |
| 10 |
Hb_001769_020 |
0.1948143688 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_000212_130 |
0.2004309482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002843_100 |
0.2014271473 |
- |
- |
- |
| 13 |
Hb_000413_170 |
0.2020826312 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |
| 14 |
Hb_179511_010 |
0.2032154048 |
- |
- |
- |
| 15 |
Hb_003581_110 |
0.2039942384 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001633_100 |
0.2053371926 |
- |
- |
PREDICTED: protein NETWORKED 3A-like [Jatropha curcas] |
| 17 |
Hb_002609_010 |
0.2078466765 |
- |
- |
PREDICTED: uncharacterized protein LOC105630866 [Jatropha curcas] |
| 18 |
Hb_006501_060 |
0.208050138 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Jatropha curcas] |
| 19 |
Hb_002073_040 |
0.208381594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_029866_130 |
0.210428175 |
- |
- |
hypothetical protein JCGZ_00035 [Jatropha curcas] |