| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
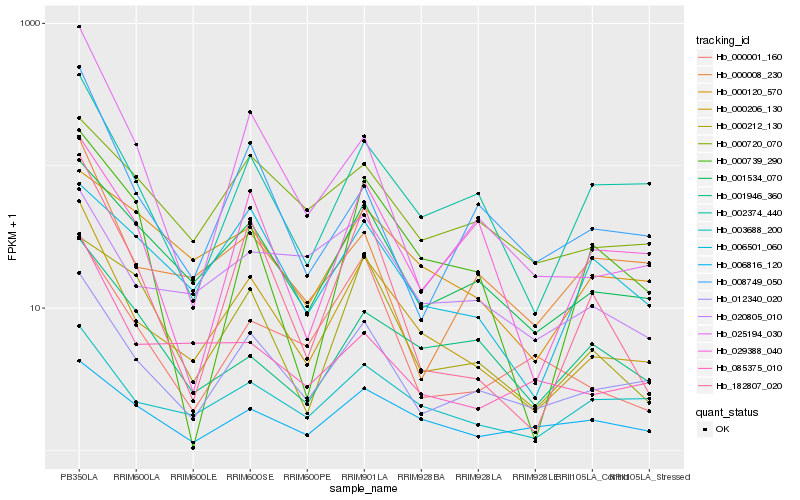
Hb_000206_130 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_012340_020 |
0.1278920205 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001534_070 |
0.1491206276 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 4 |
Hb_003688_200 |
0.1494497177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002374_440 |
0.1617344214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_182807_020 |
0.1718380668 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 7 |
Hb_006816_120 |
0.1846280956 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 8 |
Hb_085375_010 |
0.1920171672 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_029388_040 |
0.1967659392 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 10 |
Hb_020805_010 |
0.1977065807 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_008749_050 |
0.1984680054 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C4H3.01 [Jatropha curcas] |
| 12 |
Hb_025194_030 |
0.1986916995 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 13 |
Hb_000720_070 |
0.1994435454 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 14 |
Hb_000008_230 |
0.1995339445 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 15 |
Hb_000001_160 |
0.1998088528 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000212_130 |
0.1998697186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000120_570 |
0.2002254772 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 18 |
Hb_006501_060 |
0.2011611648 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Jatropha curcas] |
| 19 |
Hb_001946_360 |
0.2039684753 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 20 |
Hb_000739_290 |
0.2040182775 |
- |
- |
cytochrome P450, putative [Ricinus communis] |