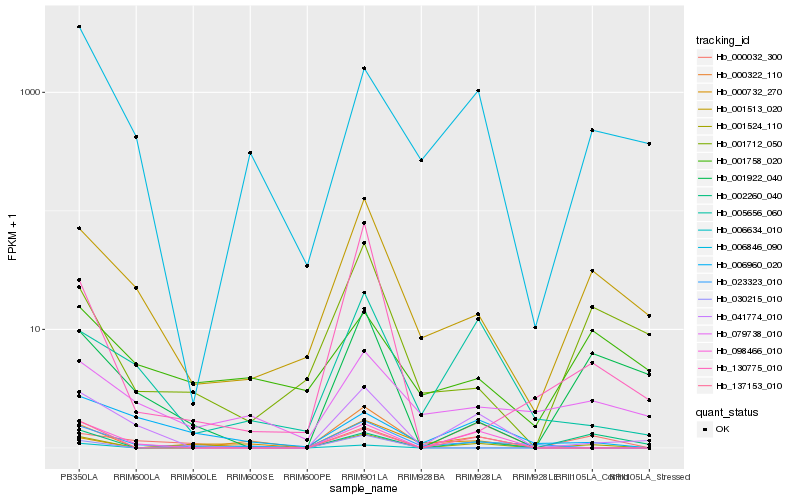
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001524_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104611877 isoform X2 [Nelumbo nucifera] |
| 2 |
Hb_000732_270 |
0.3306436327 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 3 |
Hb_000322_110 |
0.3337284411 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 3 [Jatropha curcas] |
| 4 |
Hb_137153_010 |
0.3445394243 |
- |
- |
- |
| 5 |
Hb_002260_040 |
0.3492127979 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 6 |
Hb_023323_010 |
0.3522500836 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_000032_300 |
0.3545638136 |
- |
- |
hypothetical protein JCGZ_20455 [Jatropha curcas] |
| 8 |
Hb_001513_020 |
0.3549245048 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 9 |
Hb_030215_010 |
0.3573667778 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 10 |
Hb_001712_050 |
0.3648308166 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 11 |
Hb_098466_010 |
0.3680488483 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 12 |
Hb_041774_010 |
0.3686865588 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 13 |
Hb_006960_020 |
0.372022381 |
- |
- |
PREDICTED: uncharacterized protein LOC102617255 [Citrus sinensis] |
| 14 |
Hb_006846_090 |
0.3750753024 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 15 |
Hb_079738_010 |
0.3770690291 |
- |
- |
hypothetical protein CISIN_1g036582mg, partial [Citrus sinensis] |
| 16 |
Hb_001922_040 |
0.3787022654 |
- |
- |
reticulon family protein [Populus trichocarpa] |
| 17 |
Hb_005656_060 |
0.3787051142 |
- |
- |
- |
| 18 |
Hb_006634_010 |
0.3798264689 |
- |
- |
- |
| 19 |
Hb_001758_020 |
0.3806647319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_130775_010 |
0.3827002165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |