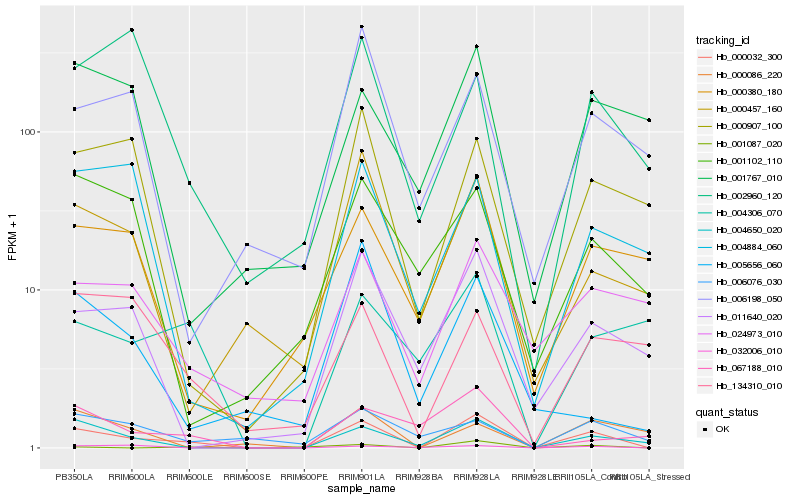
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_300 |
0.0 |
- |
- |
hypothetical protein JCGZ_20455 [Jatropha curcas] |
| 2 |
Hb_004650_020 |
0.2578364383 |
- |
- |
- |
| 3 |
Hb_011640_020 |
0.2955902262 |
- |
- |
hypothetical protein VITISV_036432 [Vitis vinifera] |
| 4 |
Hb_001087_020 |
0.3046967417 |
- |
- |
PREDICTED: phytoene synthase 2, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000457_160 |
0.3060887475 |
- |
- |
PREDICTED: uncharacterized protein LOC105633076 [Jatropha curcas] |
| 6 |
Hb_032006_010 |
0.3079790094 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 7 |
Hb_000907_100 |
0.3105343849 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0012s03770g, partial [Populus trichocarpa] |
| 8 |
Hb_000380_180 |
0.3120329612 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_024973_010 |
0.3150683112 |
- |
- |
PREDICTED: uncharacterized protein LOC105642097 isoform X2 [Jatropha curcas] |
| 10 |
Hb_067188_010 |
0.3164246828 |
- |
- |
- |
| 11 |
Hb_134310_010 |
0.3225669383 |
- |
- |
PREDICTED: ATP-dependent DNA helicase MER3 homolog [Jatropha curcas] |
| 12 |
Hb_006076_030 |
0.323988958 |
desease resistance |
Gene Name: NB-ARC |
NBS resistance protein [Hevea brasiliensis] |
| 13 |
Hb_002960_120 |
0.3260171838 |
- |
- |
PREDICTED: uncharacterized protein LOC105635318 [Jatropha curcas] |
| 14 |
Hb_004884_060 |
0.3270906779 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative Myb family transcription factor At1g14600 [Jatropha curcas] |
| 15 |
Hb_006198_050 |
0.3284436105 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_005656_060 |
0.3304011887 |
- |
- |
- |
| 17 |
Hb_000086_220 |
0.3313629687 |
- |
- |
- |
| 18 |
Hb_001102_110 |
0.3316203963 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1-like [Jatropha curcas] |
| 19 |
Hb_001767_010 |
0.3323247856 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 20 |
Hb_004306_070 |
0.332414813 |
- |
- |
hypothetical protein 0_18789_01, partial [Pinus taeda] |