| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
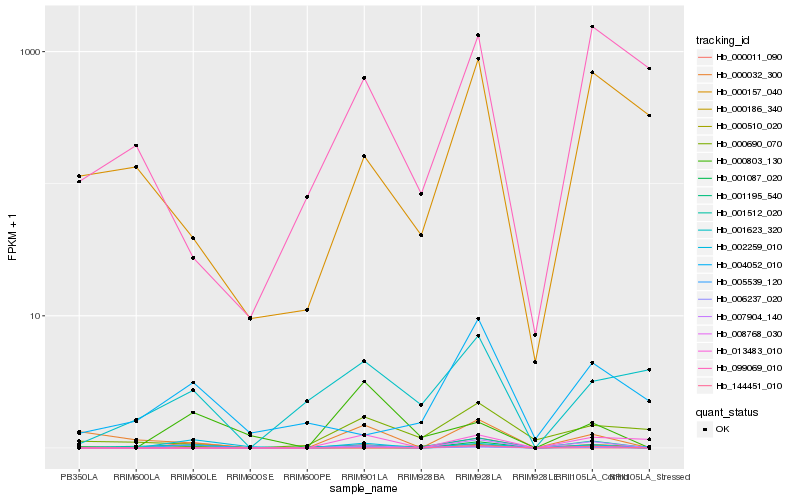
Hb_001195_540 |
0.0 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 2 |
Hb_002259_010 |
0.2987452897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001087_020 |
0.3054531504 |
- |
- |
PREDICTED: phytoene synthase 2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_001512_020 |
0.3086303364 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 5 |
Hb_007904_140 |
0.3148546392 |
- |
- |
- |
| 6 |
Hb_005539_120 |
0.3558320076 |
- |
- |
PREDICTED: probable LRR receptor-like protein kinase At1g51890 [Jatropha curcas] |
| 7 |
Hb_008768_030 |
0.3799374195 |
- |
- |
- |
| 8 |
Hb_000011_090 |
0.3888229342 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 9 |
Hb_000803_130 |
0.3994292499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000032_300 |
0.4056396334 |
- |
- |
hypothetical protein JCGZ_20455 [Jatropha curcas] |
| 11 |
Hb_006237_020 |
0.4253317463 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_004052_010 |
0.4260045405 |
- |
- |
PREDICTED: ninja-family protein AFP3-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001623_320 |
0.4308585092 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X1 [Populus euphratica] |
| 14 |
Hb_013483_010 |
0.4318984023 |
- |
- |
- |
| 15 |
Hb_000186_340 |
0.4361008864 |
- |
- |
hypothetical protein POPTR_0011s01440g [Populus trichocarpa] |
| 16 |
Hb_000690_070 |
0.4515279737 |
- |
- |
- |
| 17 |
Hb_000157_040 |
0.4576142657 |
- |
- |
PREDICTED: cycloartenol-C-24-methyltransferase [Jatropha curcas] |
| 18 |
Hb_099069_010 |
0.4578133343 |
- |
- |
S-adenosyl-methionine-sterol-C- methyltransferase [Ricinus communis] |
| 19 |
Hb_000510_020 |
0.4592998713 |
- |
- |
PREDICTED: uncharacterized protein LOC103415345 [Malus domestica] |
| 20 |
Hb_144451_010 |
0.4593009207 |
- |
- |
hypothetical protein VITISV_001808 [Vitis vinifera] |