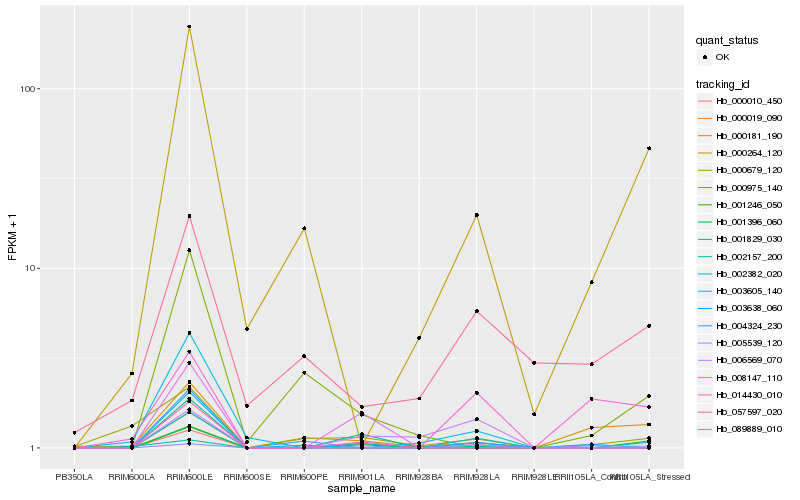
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002382_020 |
0.0 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 2 |
Hb_000010_450 |
0.3163219936 |
- |
- |
purine permease, putative [Ricinus communis] |
| 3 |
Hb_000264_120 |
0.3256849754 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 4 |
Hb_002157_200 |
0.3353921826 |
- |
- |
hypothetical protein VITISV_040170 [Vitis vinifera] |
| 5 |
Hb_000019_090 |
0.3669473478 |
- |
- |
PREDICTED: thioredoxin H4-1-like [Jatropha curcas] |
| 6 |
Hb_057597_020 |
0.3669959903 |
- |
- |
PREDICTED: anti-H(O) lectin-like [Gossypium raimondii] |
| 7 |
Hb_001396_060 |
0.3684443112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001246_050 |
0.3707024005 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 9 |
Hb_006569_070 |
0.3724951277 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_003605_140 |
0.3726613759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003638_060 |
0.3732969628 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 12 |
Hb_004324_230 |
0.3750306608 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 13 |
Hb_005539_120 |
0.3755642628 |
- |
- |
PREDICTED: probable LRR receptor-like protein kinase At1g51890 [Jatropha curcas] |
| 14 |
Hb_089889_010 |
0.3773931509 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_001829_030 |
0.3802499854 |
- |
- |
PREDICTED: uncharacterized protein LOC105119087 [Populus euphratica] |
| 16 |
Hb_000679_120 |
0.3877634221 |
- |
- |
hypothetical protein POPTR_0018s01610g [Populus trichocarpa] |
| 17 |
Hb_000181_190 |
0.3891183546 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 18 |
Hb_014430_010 |
0.3892384337 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000975_140 |
0.3912926001 |
- |
- |
lactoylglutathione lyase family protein [Populus trichocarpa] |
| 20 |
Hb_008147_110 |
0.3932361462 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |