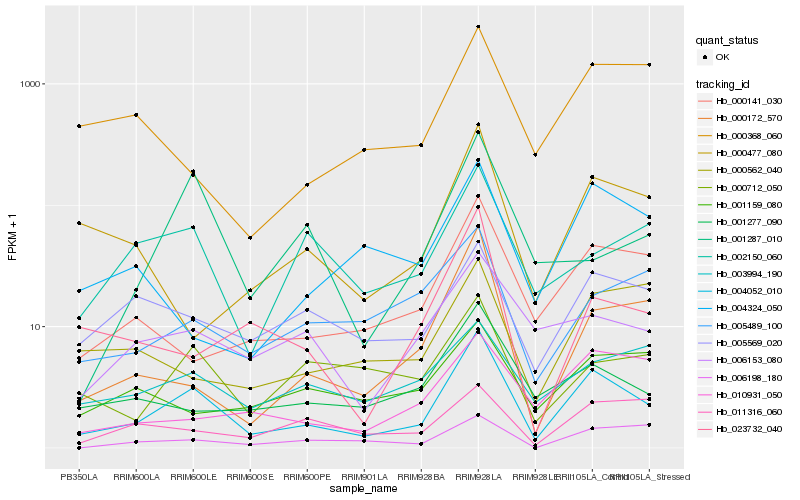
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_010 |
0.0 |
- |
- |
PREDICTED: ninja-family protein AFP3-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001277_090 |
0.2282684294 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 3 |
Hb_000141_030 |
0.2309629974 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 4 |
Hb_000172_570 |
0.2322200635 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006153_080 |
0.242939824 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |
| 6 |
Hb_005569_020 |
0.2433952176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000712_050 |
0.243587804 |
- |
- |
PREDICTED: inositol oxygenase 1-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_023732_040 |
0.2437747596 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_000477_080 |
0.2511395606 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 10 |
Hb_002150_060 |
0.2520982627 |
- |
- |
PREDICTED: arabinogalactan peptide 20-like [Jatropha curcas] |
| 11 |
Hb_005489_100 |
0.2557402423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010931_050 |
0.2567778439 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 13 |
Hb_003994_190 |
0.2615860717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006198_180 |
0.2630647117 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011316_060 |
0.2676535537 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 16 |
Hb_000562_040 |
0.2676626533 |
- |
- |
PREDICTED: auxin response factor 5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001287_010 |
0.268321918 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 18 |
Hb_004324_050 |
0.2687000491 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 19 |
Hb_000368_060 |
0.2697648289 |
- |
- |
thioredoxin H-type 6 [Hevea brasiliensis] |
| 20 |
Hb_001159_080 |
0.27313901 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |