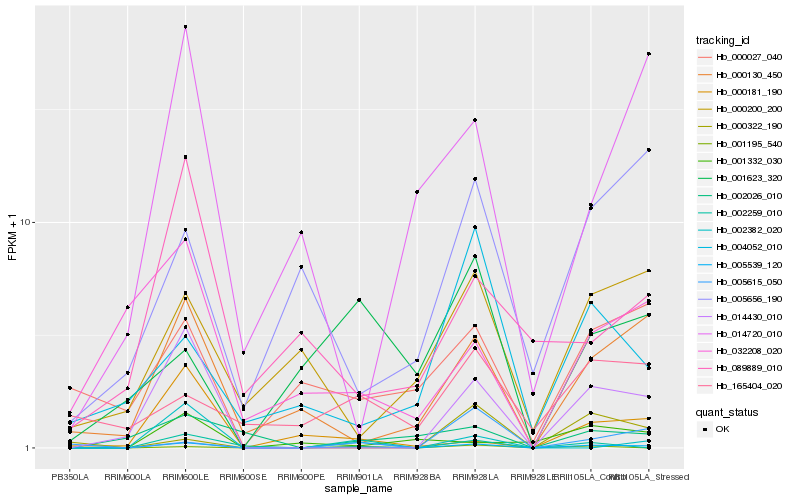
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_120 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like protein kinase At1g51890 [Jatropha curcas] |
| 2 |
Hb_014430_010 |
0.2437348286 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000130_450 |
0.3175629475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002259_010 |
0.3519896048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001195_540 |
0.3558320076 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 6 |
Hb_002382_020 |
0.3755642628 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 7 |
Hb_000181_190 |
0.3773836377 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 8 |
Hb_000027_040 |
0.3794667451 |
- |
- |
PREDICTED: uncharacterized protein LOC105636980 [Jatropha curcas] |
| 9 |
Hb_005615_050 |
0.3875125775 |
- |
- |
PREDICTED: uncharacterized protein LOC105636967 [Jatropha curcas] |
| 10 |
Hb_000200_200 |
0.3885530554 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002026_010 |
0.3948567287 |
transcription factor |
TF Family: mTERF |
hypothetical protein CICLE_v10020375mg [Citrus clementina] |
| 12 |
Hb_014720_010 |
0.3958299603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004052_010 |
0.3991899501 |
- |
- |
PREDICTED: ninja-family protein AFP3-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_089889_010 |
0.400425384 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_005656_190 |
0.4005459818 |
- |
- |
proteasome subunit beta type 6,9, putative [Ricinus communis] |
| 16 |
Hb_165404_020 |
0.4022443432 |
- |
- |
hypothetical protein JCGZ_17789 [Jatropha curcas] |
| 17 |
Hb_001623_320 |
0.4024695427 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X1 [Populus euphratica] |
| 18 |
Hb_000322_190 |
0.4028468356 |
- |
- |
- |
| 19 |
Hb_032208_020 |
0.4029690609 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 20 |
Hb_001332_030 |
0.4046651676 |
- |
- |
hypothetical protein JCGZ_21111 [Jatropha curcas] |