| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
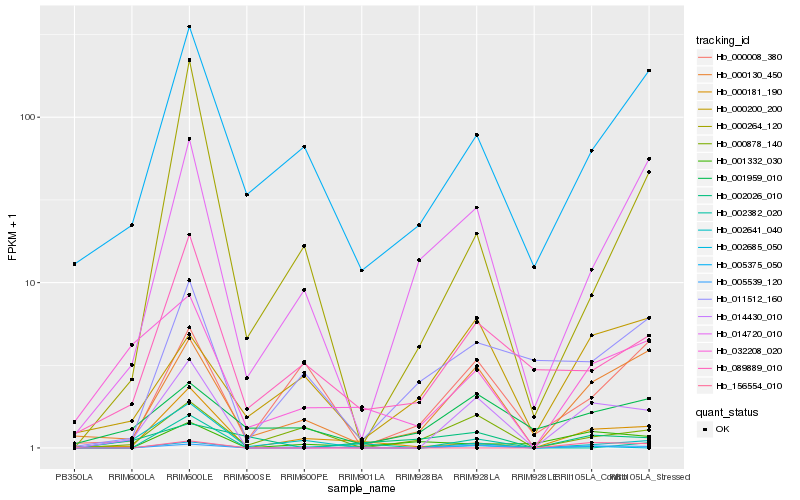
| 1 |
Hb_014430_010 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_005539_120 |
0.2437348286 |
- |
- |
PREDICTED: probable LRR receptor-like protein kinase At1g51890 [Jatropha curcas] |
| 3 |
Hb_000130_450 |
0.278875003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_014720_010 |
0.3336931436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000181_190 |
0.3405018615 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 6 |
Hb_000878_140 |
0.3478306179 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 7 |
Hb_089889_010 |
0.3501797777 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_032208_020 |
0.356752541 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 9 |
Hb_002685_050 |
0.358018645 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005375_050 |
0.3598682635 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 11 |
Hb_000264_120 |
0.3620507681 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 12 |
Hb_011512_160 |
0.3790064573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001959_010 |
0.3790123219 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 14 |
Hb_156554_010 |
0.3793235746 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 15 |
Hb_002641_040 |
0.3825601118 |
- |
- |
class III peroxidase [Populus trichocarpa] |
| 16 |
Hb_000200_200 |
0.3837817725 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000008_380 |
0.3884487086 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_002382_020 |
0.3892384337 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 19 |
Hb_001332_030 |
0.3916143133 |
- |
- |
hypothetical protein JCGZ_21111 [Jatropha curcas] |
| 20 |
Hb_002026_010 |
0.401106516 |
transcription factor |
TF Family: mTERF |
hypothetical protein CICLE_v10020375mg [Citrus clementina] |