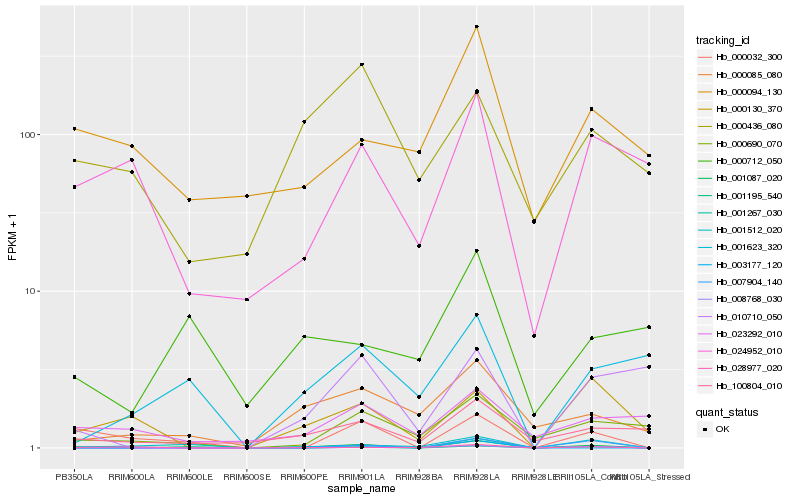
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001087_020 |
0.0 |
- |
- |
PREDICTED: phytoene synthase 2, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000032_300 |
0.3046967417 |
- |
- |
hypothetical protein JCGZ_20455 [Jatropha curcas] |
| 3 |
Hb_001195_540 |
0.3054531504 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 4 |
Hb_001512_020 |
0.3138182868 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 5 |
Hb_007904_140 |
0.3522250924 |
- |
- |
- |
| 6 |
Hb_100804_010 |
0.359801714 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 7 |
Hb_000085_080 |
0.3666710062 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_000690_070 |
0.3792462394 |
- |
- |
- |
| 9 |
Hb_001267_030 |
0.3857182439 |
- |
- |
hypothetical protein VITISV_041979 [Vitis vinifera] |
| 10 |
Hb_028977_020 |
0.3881007041 |
- |
- |
hypothetical protein JCGZ_21319 [Jatropha curcas] |
| 11 |
Hb_000712_050 |
0.3944459599 |
- |
- |
PREDICTED: inositol oxygenase 1-like isoform X1 [Citrus sinensis] |
| 12 |
Hb_000094_130 |
0.3947368512 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 13 |
Hb_003177_120 |
0.3964461592 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Populus euphratica] |
| 14 |
Hb_001623_320 |
0.3995729021 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X1 [Populus euphratica] |
| 15 |
Hb_010710_050 |
0.4007350005 |
- |
- |
- |
| 16 |
Hb_000130_370 |
0.4031528191 |
- |
- |
PREDICTED: uncharacterized protein LOC105635737 [Jatropha curcas] |
| 17 |
Hb_024952_010 |
0.4057030535 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 18 |
Hb_008768_030 |
0.4060099011 |
- |
- |
- |
| 19 |
Hb_023292_010 |
0.4069218102 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 20 |
Hb_000436_080 |
0.412218363 |
- |
- |
unnamed protein product [Coffea canephora] |