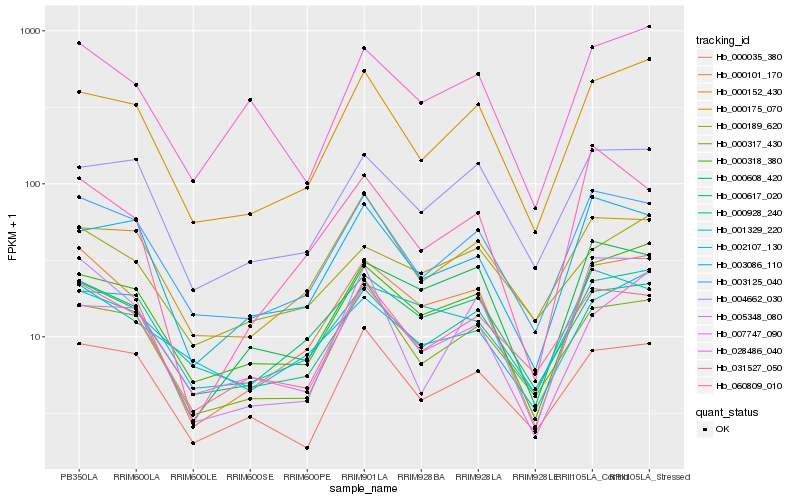
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000101_170 |
0.0 |
- |
- |
- |
| 2 |
Hb_000318_380 |
0.1014303123 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 3-like [Jatropha curcas] |
| 3 |
Hb_000617_020 |
0.106300672 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000152_430 |
0.1112604939 |
- |
- |
PREDICTED: 40S ribosomal protein S20-1 [Jatropha curcas] |
| 5 |
Hb_003125_040 |
0.1115569264 |
- |
- |
PREDICTED: uncharacterized protein LOC105640201 isoform X1 [Jatropha curcas] |
| 6 |
Hb_060809_010 |
0.1218443091 |
- |
- |
PREDICTED: uncharacterized protein LOC105163892 [Sesamum indicum] |
| 7 |
Hb_000317_430 |
0.1286253284 |
transcription factor |
TF Family: SET |
PREDICTED: ribosomal lysine N-methyltransferase 3 [Jatropha curcas] |
| 8 |
Hb_000928_240 |
0.1311523666 |
- |
- |
PREDICTED: uncharacterized protein LOC105632515 [Jatropha curcas] |
| 9 |
Hb_028486_040 |
0.1323124519 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 10 |
Hb_000608_420 |
0.1331165314 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 11 |
Hb_000189_620 |
0.1340600348 |
- |
- |
hypothetical protein JCGZ_23450 [Jatropha curcas] |
| 12 |
Hb_004662_030 |
0.1342417466 |
- |
- |
hypothetical protein PRUPE_ppa012487mg [Prunus persica] |
| 13 |
Hb_002107_130 |
0.1348098665 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_031527_050 |
0.134884737 |
- |
- |
PREDICTED: universal stress protein A-like protein [Populus euphratica] |
| 15 |
Hb_001329_220 |
0.1358424297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005348_080 |
0.1371140958 |
- |
- |
PREDICTED: oligouridylate-binding protein 1-like [Jatropha curcas] |
| 17 |
Hb_003086_110 |
0.1374684203 |
- |
- |
PREDICTED: dihydroneopterin aldolase-like isoform X2 [Prunus mume] |
| 18 |
Hb_000035_380 |
0.1375997586 |
- |
- |
- |
| 19 |
Hb_007747_090 |
0.1381196157 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 20 |
Hb_000175_070 |
0.1399273089 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 11 isoform X1 [Jatropha curcas] |