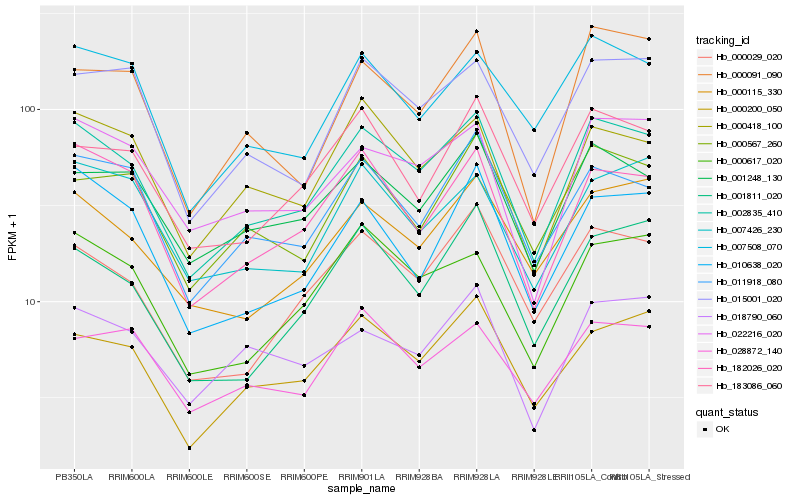
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_410 |
0.0 |
- |
- |
clathrin coat assembly protein ap-1, putative [Ricinus communis] |
| 2 |
Hb_182026_020 |
0.0793461107 |
- |
- |
PREDICTED: SUN domain-containing protein 3-like [Jatropha curcas] |
| 3 |
Hb_022216_020 |
0.0804007253 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_000200_050 |
0.0806426724 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001248_130 |
0.0806934996 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000029_020 |
0.0839073875 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 7 |
Hb_000418_100 |
0.0843351319 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 8 |
Hb_000617_020 |
0.0845313737 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000115_330 |
0.0852213291 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 10 |
Hb_015001_020 |
0.0861619043 |
- |
- |
PREDICTED: la-related protein 6B [Jatropha curcas] |
| 11 |
Hb_011918_080 |
0.087062126 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007508_070 |
0.0885512717 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 13 |
Hb_183086_060 |
0.0918942721 |
desease resistance |
Gene Name: AAA |
PREDICTED: ATPase family AAA domain-containing protein 1-B [Jatropha curcas] |
| 14 |
Hb_000567_260 |
0.0927260496 |
- |
- |
BnaA09g41440D [Brassica napus] |
| 15 |
Hb_007426_230 |
0.0958937716 |
- |
- |
hypothetical protein CICLE_v10017932mg [Citrus clementina] |
| 16 |
Hb_010638_020 |
0.0960947537 |
- |
- |
PREDICTED: uncharacterized protein LOC105638189 isoform X3 [Jatropha curcas] |
| 17 |
Hb_028872_140 |
0.0963667417 |
- |
- |
PREDICTED: uncharacterized protein LOC105644767 isoform X1 [Jatropha curcas] |
| 18 |
Hb_018790_060 |
0.0971512477 |
- |
- |
hypothetical protein JCGZ_21628 [Jatropha curcas] |
| 19 |
Hb_001811_020 |
0.0974692543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000091_090 |
0.0980345801 |
- |
- |
hypothetical protein JCGZ_09031 [Jatropha curcas] |