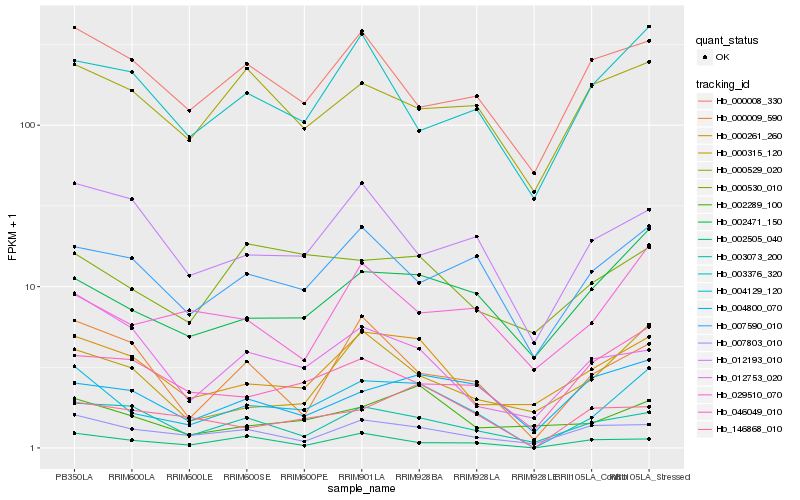
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004129_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_000261_260 |
0.179688101 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: HBS1-like protein [Populus euphratica] |
| 3 |
Hb_002471_150 |
0.1849002825 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 4 |
Hb_007803_010 |
0.1931237205 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 5 |
Hb_007590_010 |
0.1949150052 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_003073_200 |
0.1991751582 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 7 |
Hb_000009_590 |
0.2002807527 |
- |
- |
PREDICTED: prolyl endopeptidase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_046049_010 |
0.2042203827 |
- |
- |
PREDICTED: far upstream element-binding protein 2 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000315_120 |
0.2052728036 |
- |
- |
- |
| 10 |
Hb_012193_010 |
0.2055070634 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_002289_100 |
0.20666589 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 12 |
Hb_146868_010 |
0.2068403121 |
- |
- |
- |
| 13 |
Hb_000008_330 |
0.2069051216 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 14 |
Hb_003376_320 |
0.2077897058 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 15 |
Hb_002505_040 |
0.2094430849 |
- |
- |
- |
| 16 |
Hb_000529_020 |
0.2101030577 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 17 |
Hb_012753_020 |
0.2116814676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_029510_070 |
0.2119119796 |
- |
- |
Cytochrome c-type biogenesis protein cycL precursor, putative [Ricinus communis] |
| 19 |
Hb_004800_070 |
0.2124200793 |
- |
- |
- |
| 20 |
Hb_000530_010 |
0.2128860088 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |