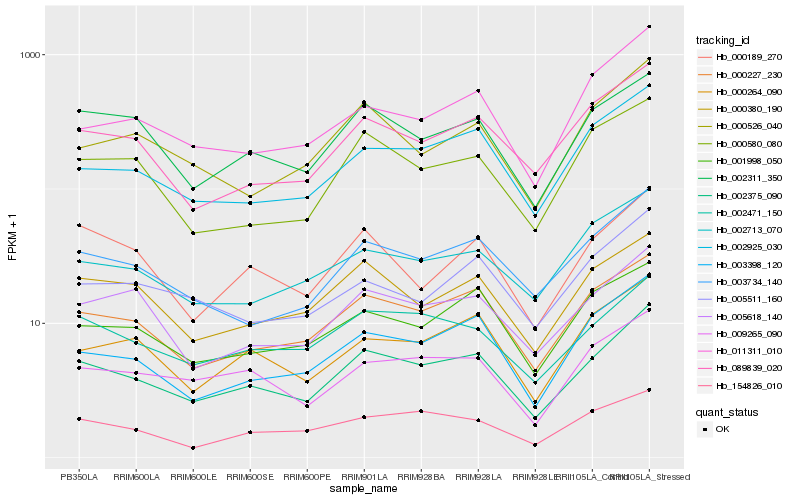
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002375_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000227_230 |
0.0744484767 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 3 |
Hb_002471_150 |
0.0868450127 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 4 |
Hb_003734_140 |
0.0869208485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002925_030 |
0.0874465146 |
- |
- |
40S ribosomal protein S16B [Hevea brasiliensis] |
| 6 |
Hb_089839_020 |
0.0917564121 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 7 |
Hb_003398_120 |
0.09201386 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 8 |
Hb_009265_090 |
0.0925949282 |
- |
- |
PREDICTED: uncharacterized protein LOC105649618 [Jatropha curcas] |
| 9 |
Hb_002311_350 |
0.0948757596 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 10 |
Hb_000264_090 |
0.0962056179 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 11 |
Hb_005618_140 |
0.096358856 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 12 |
Hb_000380_190 |
0.0971087474 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 13 |
Hb_001998_050 |
0.1002085473 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_002713_070 |
0.1023747027 |
- |
- |
hypothetical protein POPTR_0002s21620g [Populus trichocarpa] |
| 15 |
Hb_000189_270 |
0.1052562775 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 16 |
Hb_000580_080 |
0.1063093474 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 17 |
Hb_154826_010 |
0.1072333625 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_000526_040 |
0.1074618864 |
- |
- |
60S ribosomal protein L6, putative [Ricinus communis] |
| 19 |
Hb_005511_160 |
0.108924852 |
- |
- |
OB-fold nucleic acid binding domain-containing protein [Theobroma cacao] |
| 20 |
Hb_011311_010 |
0.1095533801 |
- |
- |
60S ribosomal protein L27C [Hevea brasiliensis] |