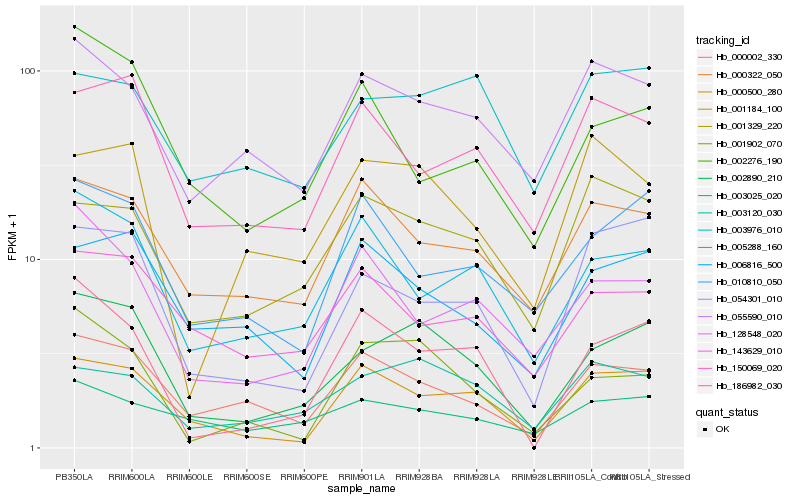
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_210 |
0.0 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 2 |
Hb_001902_070 |
0.1582941597 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_003120_030 |
0.1642162253 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 4 |
Hb_000002_330 |
0.1643264072 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 5 |
Hb_000500_280 |
0.1651943507 |
- |
- |
Brassinosteroid-regulated protein BRU1 precursor, putative [Ricinus communis] |
| 6 |
Hb_054301_010 |
0.1653476372 |
- |
- |
hypothetical protein EUTSA_v10006737mg [Eutrema salsugineum] |
| 7 |
Hb_186982_030 |
0.1699656586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005288_160 |
0.1783619121 |
- |
- |
PREDICTED: uncharacterized protein LOC105647746 isoform X1 [Jatropha curcas] |
| 9 |
Hb_055590_010 |
0.1824634255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006816_500 |
0.1838539835 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 11 |
Hb_003025_020 |
0.1844860904 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_010810_050 |
0.1862549956 |
- |
- |
hypothetical protein [Cleome spinosa] |
| 13 |
Hb_001184_100 |
0.1870958672 |
- |
- |
PREDICTED: zinc finger protein 4-like [Populus euphratica] |
| 14 |
Hb_000322_050 |
0.1874312571 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 15 |
Hb_150069_020 |
0.1887916143 |
- |
- |
PREDICTED: ribosome biogenesis regulatory protein homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_001329_220 |
0.1906980279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003976_010 |
0.1908438961 |
- |
- |
methionine aminopeptidase, putative [Ricinus communis] |
| 18 |
Hb_143629_010 |
0.192552657 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 19 |
Hb_128548_020 |
0.1929063952 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 20 |
Hb_002276_190 |
0.1937851537 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |