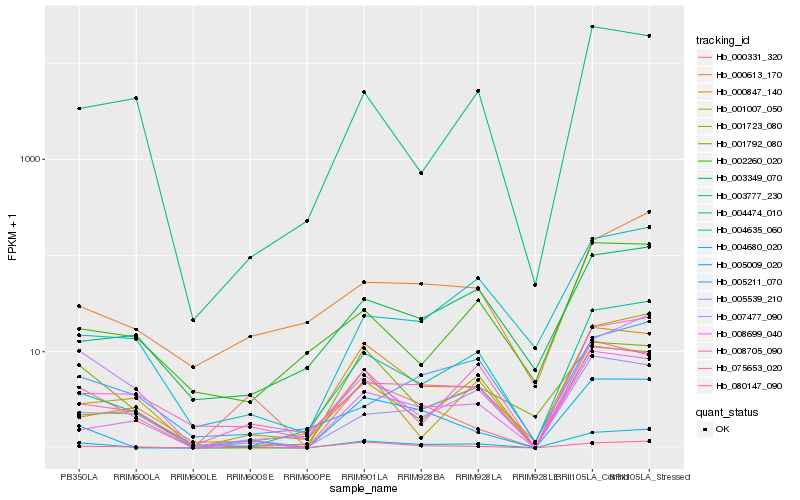
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004680_020 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105789475 [Gossypium raimondii] |
| 2 |
Hb_004474_010 |
0.1456366761 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_005009_020 |
0.1558440019 |
- |
- |
- |
| 4 |
Hb_008699_040 |
0.1644879508 |
- |
- |
- |
| 5 |
Hb_075653_020 |
0.1857867195 |
- |
- |
- |
| 6 |
Hb_008705_090 |
0.190373853 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2-like [Jatropha curcas] |
| 7 |
Hb_001723_080 |
0.1960762581 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 8 |
Hb_007477_090 |
0.1966913425 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 9 |
Hb_001007_050 |
0.1996985853 |
- |
- |
hypothetical protein MIMGU_mgv1a0196712mg, partial [Erythranthe guttata] |
| 10 |
Hb_000613_170 |
0.2003159861 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 11 |
Hb_003349_070 |
0.2027361396 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 12 |
Hb_000847_140 |
0.2054815427 |
- |
- |
- |
| 13 |
Hb_005539_210 |
0.2079903523 |
- |
- |
- |
| 14 |
Hb_004635_060 |
0.2101912004 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |
| 15 |
Hb_080147_090 |
0.2123199503 |
- |
- |
hypothetical protein JCGZ_07271 [Jatropha curcas] |
| 16 |
Hb_003777_230 |
0.2155557383 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 17 |
Hb_002260_020 |
0.2181789682 |
- |
- |
- |
| 18 |
Hb_001792_080 |
0.2244442875 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_000331_320 |
0.2249358832 |
- |
- |
- |
| 20 |
Hb_005211_070 |
0.2267818135 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |