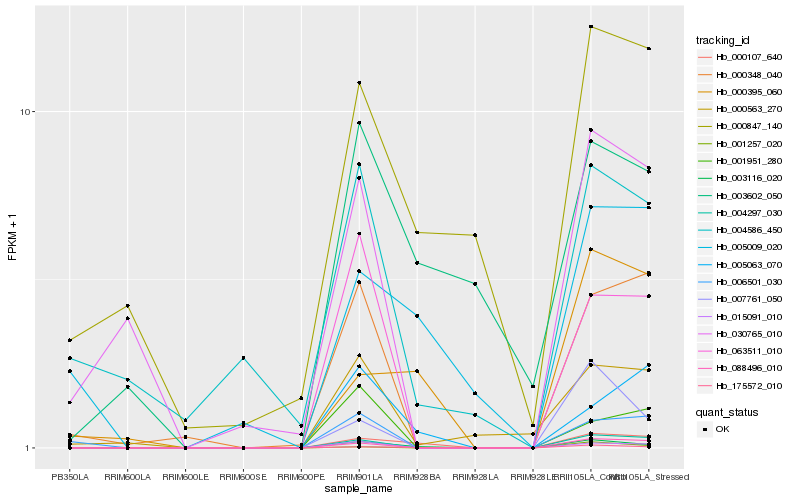
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003116_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 2 |
Hb_175572_010 |
0.1210837725 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 3 |
Hb_000107_640 |
0.1523538622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_015091_010 |
0.2237047439 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 5 |
Hb_004297_030 |
0.2361631382 |
- |
- |
PREDICTED: uncharacterized protein LOC105633198 [Jatropha curcas] |
| 6 |
Hb_088496_010 |
0.2493039535 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_063511_010 |
0.2545367599 |
- |
- |
DNA-directed RNA polymerase II subunit RPB4 [Gossypium arboreum] |
| 8 |
Hb_007761_050 |
0.2646223355 |
- |
- |
calcineurin-like protein [Hevea brasiliensis] |
| 9 |
Hb_003602_050 |
0.2771075924 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_005063_070 |
0.2793422333 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 11 |
Hb_000395_060 |
0.28946781 |
- |
- |
- |
| 12 |
Hb_000563_270 |
0.2927140204 |
- |
- |
- |
| 13 |
Hb_000348_040 |
0.2936007508 |
- |
- |
- |
| 14 |
Hb_001257_020 |
0.2990065832 |
- |
- |
- |
| 15 |
Hb_000847_140 |
0.2997119048 |
- |
- |
- |
| 16 |
Hb_005009_020 |
0.3042264677 |
- |
- |
- |
| 17 |
Hb_006501_030 |
0.30562034 |
- |
- |
hypothetical protein POPTR_0006s17720g [Populus trichocarpa] |
| 18 |
Hb_001951_280 |
0.3087258495 |
- |
- |
- |
| 19 |
Hb_030765_010 |
0.3116054781 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter G family member 11-like isoform X1 [Solanum lycopersicum] |
| 20 |
Hb_004586_450 |
0.311754042 |
- |
- |
- |