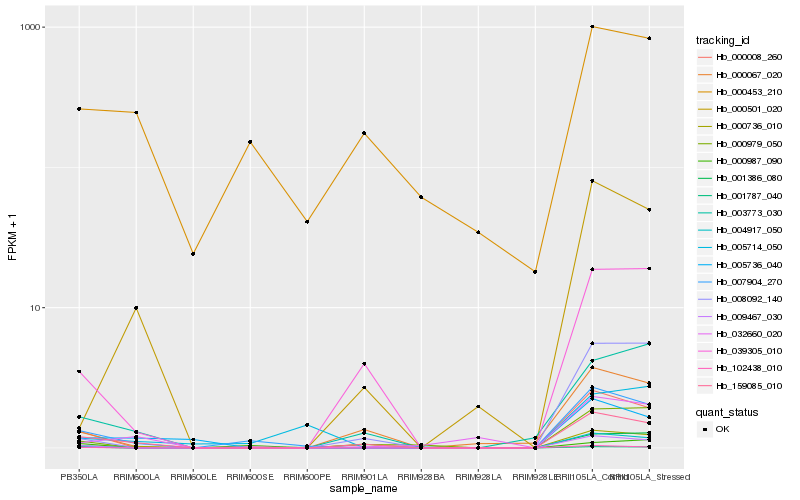
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007904_270 |
0.0 |
- |
- |
- |
| 2 |
Hb_000008_260 |
0.1893381918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009467_030 |
0.1897587174 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 4 |
Hb_102438_010 |
0.2086186569 |
- |
- |
- |
| 5 |
Hb_159085_010 |
0.2181066093 |
- |
- |
- |
| 6 |
Hb_004917_050 |
0.2272458332 |
- |
- |
- |
| 7 |
Hb_000736_010 |
0.2550855637 |
- |
- |
PREDICTED: uncharacterized protein LOC105762083 [Gossypium raimondii] |
| 8 |
Hb_001386_080 |
0.2571000816 |
- |
- |
hypothetical protein JCGZ_06052 [Jatropha curcas] |
| 9 |
Hb_005736_040 |
0.2579653319 |
- |
- |
- |
| 10 |
Hb_005714_050 |
0.275034787 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_1461s00200g [Populus trichocarpa] |
| 11 |
Hb_039305_010 |
0.2757767947 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 12 |
Hb_000979_050 |
0.2768668213 |
- |
- |
- |
| 13 |
Hb_003773_030 |
0.2781300774 |
- |
- |
- |
| 14 |
Hb_008092_140 |
0.2829792366 |
- |
- |
PREDICTED: uncharacterized protein LOC103636948 [Zea mays] |
| 15 |
Hb_000067_020 |
0.2833582181 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 16 |
Hb_000453_210 |
0.2844908735 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 17 |
Hb_000987_090 |
0.2849746199 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 18 |
Hb_032660_020 |
0.2853782487 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 19 |
Hb_000501_020 |
0.2892540733 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 20 |
Hb_001787_040 |
0.2911411038 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |