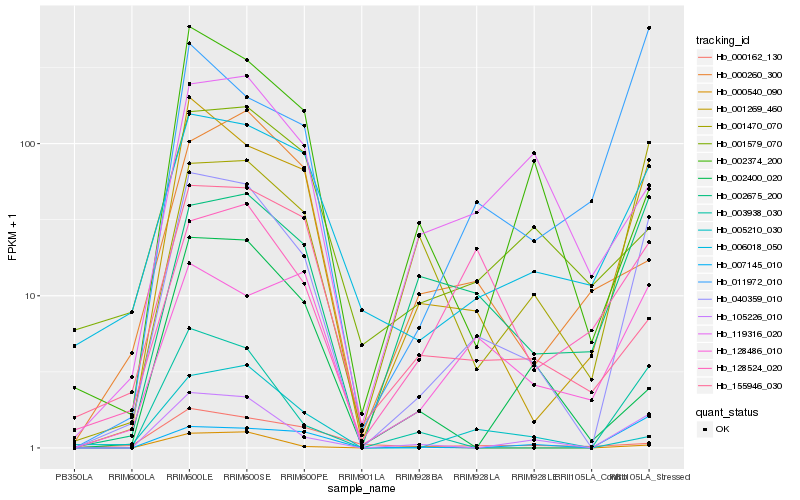
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_040359_010 |
0.0 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 2 |
Hb_001269_460 |
0.1543367352 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 3 |
Hb_105226_010 |
0.158027757 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 4 |
Hb_006018_050 |
0.1950465216 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 5 |
Hb_003938_030 |
0.19770399 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 6 |
Hb_002675_200 |
0.2330536567 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 7 |
Hb_001470_070 |
0.243979895 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_155946_030 |
0.2505464806 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 9 |
Hb_007145_010 |
0.2507976833 |
- |
- |
ammonium transporter 3.1-like protein [Camellia sinensis] |
| 10 |
Hb_005210_030 |
0.2549543019 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 11 |
Hb_128486_010 |
0.255764396 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 12 |
Hb_000540_090 |
0.2581260876 |
- |
- |
PREDICTED: chitinase 2-like [Jatropha curcas] |
| 13 |
Hb_119316_020 |
0.2588117965 |
- |
- |
- |
| 14 |
Hb_002374_200 |
0.2588594131 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 15 |
Hb_001579_070 |
0.2590996531 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 16 |
Hb_011972_010 |
0.2611780803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_128524_020 |
0.2652078419 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000260_300 |
0.273725258 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 19 |
Hb_002400_020 |
0.274437292 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 20 |
Hb_000162_130 |
0.27533046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |