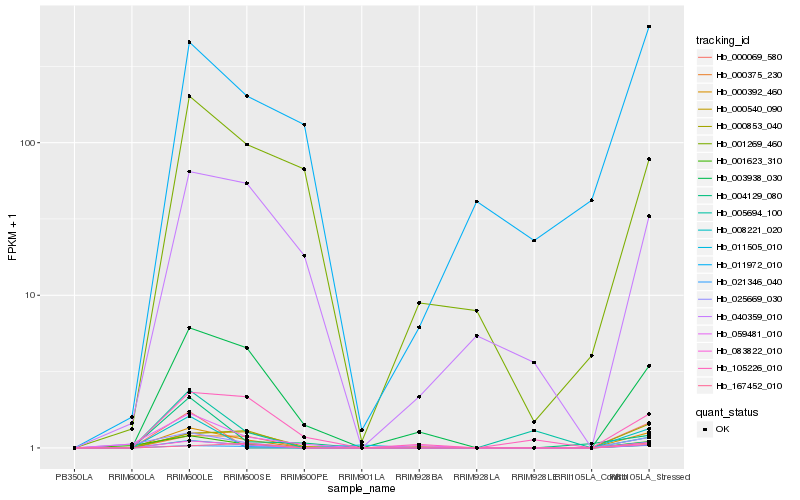
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083822_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_000392_460 |
0.1034694441 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 3 |
Hb_021346_040 |
0.1651338257 |
- |
- |
hypothetical protein JCGZ_26751 [Jatropha curcas] |
| 4 |
Hb_003938_030 |
0.1928970651 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 5 |
Hb_000375_230 |
0.2033393341 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 6 |
Hb_001623_310 |
0.2271164925 |
- |
- |
hypothetical protein CISIN_1g042596mg [Citrus sinensis] |
| 7 |
Hb_000853_040 |
0.2521124888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_025669_030 |
0.2532875168 |
- |
- |
PREDICTED: uncharacterized protein LOC105649301 [Jatropha curcas] |
| 9 |
Hb_105226_010 |
0.266968153 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 10 |
Hb_011505_010 |
0.3032112938 |
- |
- |
- |
| 11 |
Hb_040359_010 |
0.3042763067 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 12 |
Hb_001269_460 |
0.3178891753 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 13 |
Hb_000540_090 |
0.3179897356 |
- |
- |
PREDICTED: chitinase 2-like [Jatropha curcas] |
| 14 |
Hb_167452_010 |
0.3187828902 |
- |
- |
hypothetical protein JCGZ_03682 [Jatropha curcas] |
| 15 |
Hb_004129_080 |
0.3214767477 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 16 |
Hb_059481_010 |
0.3265270522 |
- |
- |
hypothetical protein POPTR_0008s06860g [Populus trichocarpa] |
| 17 |
Hb_005694_100 |
0.3405645533 |
- |
- |
PREDICTED: calcium-binding protein KIC-like [Populus euphratica] |
| 18 |
Hb_011972_010 |
0.3509109683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008221_020 |
0.3525146682 |
- |
- |
hypothetical protein JCGZ_15014 [Jatropha curcas] |
| 20 |
Hb_000069_580 |
0.356023715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |