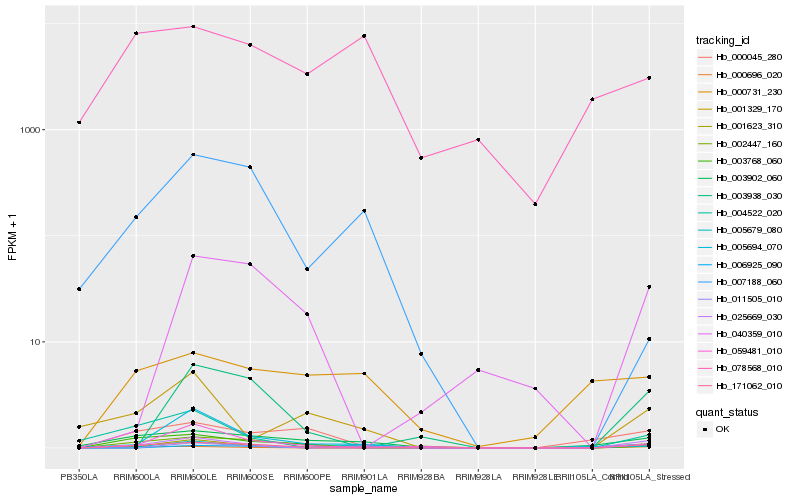
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171062_010 |
0.0 |
- |
- |
hypothetical protein VITISV_036059 [Vitis vinifera] |
| 2 |
Hb_003902_060 |
0.2365297987 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 3 |
Hb_025669_030 |
0.269748567 |
- |
- |
PREDICTED: uncharacterized protein LOC105649301 [Jatropha curcas] |
| 4 |
Hb_001623_310 |
0.2750704692 |
- |
- |
hypothetical protein CISIN_1g042596mg [Citrus sinensis] |
| 5 |
Hb_002447_160 |
0.3012532613 |
- |
- |
- |
| 6 |
Hb_000045_280 |
0.307639982 |
- |
- |
hypothetical protein JCGZ_04076 [Jatropha curcas] |
| 7 |
Hb_003768_060 |
0.3112302478 |
- |
- |
PREDICTED: transcription initiation factor IIB-like [Jatropha curcas] |
| 8 |
Hb_004522_020 |
0.3139596903 |
- |
- |
PREDICTED: uncharacterized protein LOC105646836 [Jatropha curcas] |
| 9 |
Hb_006925_090 |
0.314427423 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 10 |
Hb_005679_080 |
0.3249786342 |
- |
- |
PREDICTED: PH-interacting protein-like [Populus euphratica] |
| 11 |
Hb_007188_060 |
0.3319752861 |
- |
- |
- |
| 12 |
Hb_059481_010 |
0.3333218017 |
- |
- |
hypothetical protein POPTR_0008s06860g [Populus trichocarpa] |
| 13 |
Hb_000731_230 |
0.3344299379 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 14 |
Hb_003938_030 |
0.3445507414 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 15 |
Hb_078568_010 |
0.3475416926 |
- |
- |
hypothetical protein PRUPE_ppa013245mg [Prunus persica] |
| 16 |
Hb_011505_010 |
0.3475516952 |
- |
- |
- |
| 17 |
Hb_005694_070 |
0.3519154106 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_001329_170 |
0.3553253393 |
- |
- |
Chlorophyll a-b binding protein, chloroplastic [Aegilops tauschii] |
| 19 |
Hb_040359_010 |
0.357094597 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 20 |
Hb_000696_020 |
0.3582914975 |
- |
- |
hypothetical protein JCGZ_25344 [Jatropha curcas] |