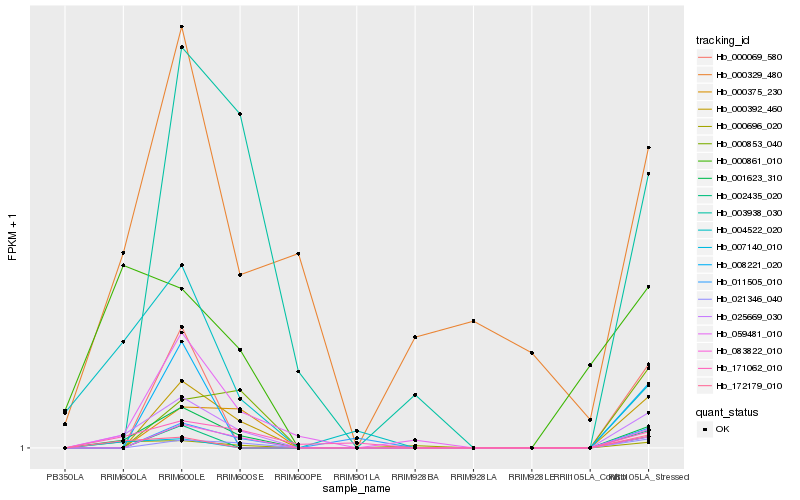
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025669_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649301 [Jatropha curcas] |
| 2 |
Hb_001623_310 |
0.0544542427 |
- |
- |
hypothetical protein CISIN_1g042596mg [Citrus sinensis] |
| 3 |
Hb_000392_460 |
0.2349541354 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 4 |
Hb_083822_010 |
0.2532875168 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_021346_040 |
0.2644272198 |
- |
- |
hypothetical protein JCGZ_26751 [Jatropha curcas] |
| 6 |
Hb_171062_010 |
0.269748567 |
- |
- |
hypothetical protein VITISV_036059 [Vitis vinifera] |
| 7 |
Hb_000696_020 |
0.2951495807 |
- |
- |
hypothetical protein JCGZ_25344 [Jatropha curcas] |
| 8 |
Hb_004522_020 |
0.3091346413 |
- |
- |
PREDICTED: uncharacterized protein LOC105646836 [Jatropha curcas] |
| 9 |
Hb_172179_010 |
0.3154984976 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 10 |
Hb_007140_010 |
0.315746952 |
- |
- |
- |
| 11 |
Hb_000853_040 |
0.3270050502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_059481_010 |
0.3271775696 |
- |
- |
hypothetical protein POPTR_0008s06860g [Populus trichocarpa] |
| 13 |
Hb_003938_030 |
0.330145343 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 14 |
Hb_000861_010 |
0.3595354645 |
- |
- |
- |
| 15 |
Hb_011505_010 |
0.3596435634 |
- |
- |
- |
| 16 |
Hb_000069_580 |
0.3676213979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002435_020 |
0.3683172874 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 18 |
Hb_008221_020 |
0.3694048809 |
- |
- |
hypothetical protein JCGZ_15014 [Jatropha curcas] |
| 19 |
Hb_000375_230 |
0.3709684503 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 20 |
Hb_000329_480 |
0.372106087 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 4 [Jatropha curcas] |