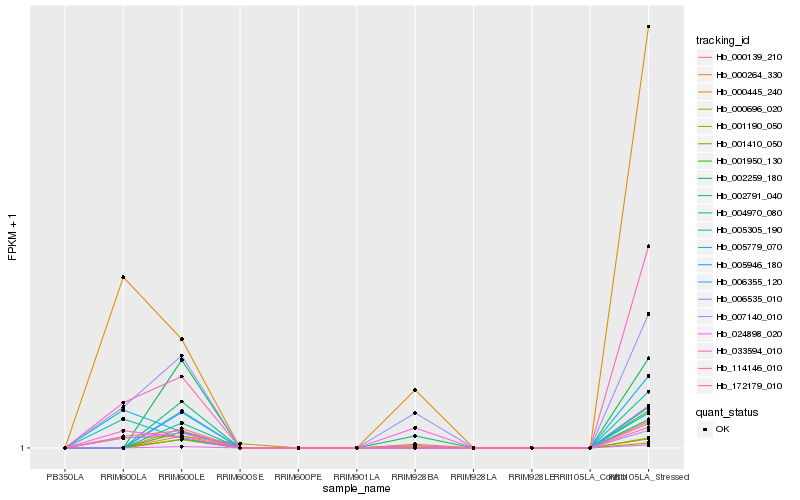
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006535_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000445_240 |
0.2433372757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_172179_010 |
0.2580558326 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 4 |
Hb_007140_010 |
0.2582441762 |
- |
- |
- |
| 5 |
Hb_033594_010 |
0.26570459 |
- |
- |
- |
| 6 |
Hb_114146_010 |
0.2739865745 |
- |
- |
hypothetical protein JCGZ_15608 [Jatropha curcas] |
| 7 |
Hb_002791_040 |
0.3122461968 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 8 |
Hb_001190_050 |
0.3128979313 |
- |
- |
hypothetical protein JCGZ_20709 [Jatropha curcas] |
| 9 |
Hb_000696_020 |
0.3173025885 |
- |
- |
hypothetical protein JCGZ_25344 [Jatropha curcas] |
| 10 |
Hb_024898_020 |
0.3498270711 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_005305_190 |
0.3593172356 |
transcription factor |
TF Family: NAC |
- |
| 12 |
Hb_001410_050 |
0.3595737361 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 13 |
Hb_005946_180 |
0.3598920888 |
- |
- |
PREDICTED: uncharacterized protein LOC100818788 [Glycine max] |
| 14 |
Hb_006355_120 |
0.3603759841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000139_210 |
0.3613418973 |
- |
- |
- |
| 16 |
Hb_000264_330 |
0.3618201783 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 17 |
Hb_005779_070 |
0.3621255777 |
- |
- |
- |
| 18 |
Hb_004970_080 |
0.3635551643 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 19 |
Hb_002259_180 |
0.3637704453 |
- |
- |
- |
| 20 |
Hb_001950_130 |
0.3640476842 |
- |
- |
- |