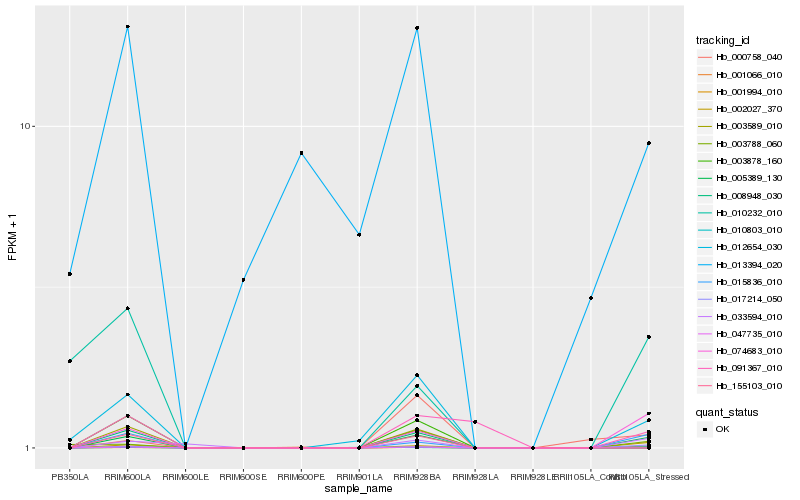
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005389_130 |
0.0 |
- |
- |
hypothetical protein CICLE_v10009852mg [Citrus clementina] |
| 2 |
Hb_001994_010 |
0.1165133521 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_074683_010 |
0.1791818522 |
- |
- |
- |
| 4 |
Hb_002027_370 |
0.195638537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_017214_050 |
0.1965721982 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_015836_010 |
0.1975429655 |
- |
- |
- |
| 7 |
Hb_012654_030 |
0.2474374055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_033594_010 |
0.2866248455 |
- |
- |
- |
| 9 |
Hb_003788_060 |
0.3407362956 |
- |
- |
PREDICTED: protein MKS1-like [Populus euphratica] |
| 10 |
Hb_001066_010 |
0.3415916278 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_091367_010 |
0.3782950941 |
- |
- |
hypothetical protein JCGZ_18029 [Jatropha curcas] |
| 12 |
Hb_010232_010 |
0.3811609676 |
- |
- |
- |
| 13 |
Hb_010803_010 |
0.3824174684 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 14 |
Hb_000758_040 |
0.3973478089 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 15 |
Hb_013394_020 |
0.4099623222 |
- |
- |
PREDICTED: putative laccase-9 [Vitis vinifera] |
| 16 |
Hb_047735_010 |
0.4118493363 |
- |
- |
DNA/RNA polymerases superfamily protein, putative [Theobroma cacao] |
| 17 |
Hb_155103_010 |
0.414412009 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_003589_010 |
0.4147612367 |
- |
- |
- |
| 19 |
Hb_008948_030 |
0.4149986864 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 20 |
Hb_003878_160 |
0.415464456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |