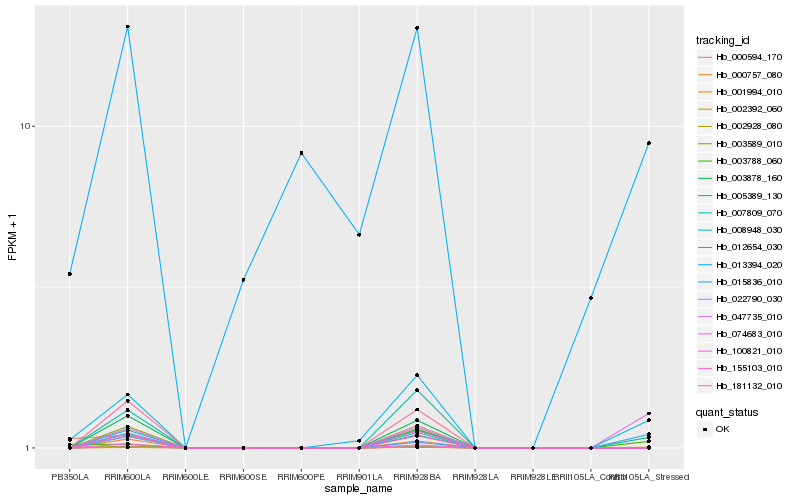
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012654_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005389_130 |
0.2474374055 |
- |
- |
hypothetical protein CICLE_v10009852mg [Citrus clementina] |
| 3 |
Hb_003788_060 |
0.2694627037 |
- |
- |
PREDICTED: protein MKS1-like [Populus euphratica] |
| 4 |
Hb_001994_010 |
0.3157298242 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_013394_020 |
0.3236899523 |
- |
- |
PREDICTED: putative laccase-9 [Vitis vinifera] |
| 6 |
Hb_007809_070 |
0.3504098942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_022790_030 |
0.3520129593 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 8 |
Hb_047735_010 |
0.3531014924 |
- |
- |
DNA/RNA polymerases superfamily protein, putative [Theobroma cacao] |
| 9 |
Hb_155103_010 |
0.3637239893 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 10 |
Hb_003589_010 |
0.364594489 |
- |
- |
- |
| 11 |
Hb_008948_030 |
0.365170281 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 12 |
Hb_003878_160 |
0.366267751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000594_170 |
0.3701622379 |
- |
- |
- |
| 14 |
Hb_181132_010 |
0.3704938663 |
- |
- |
- |
| 15 |
Hb_000757_080 |
0.3715767665 |
- |
- |
vacuolar ATP synthase subunit G plant, putative [Ricinus communis] |
| 16 |
Hb_074683_010 |
0.3753517057 |
- |
- |
- |
| 17 |
Hb_015836_010 |
0.3753777675 |
- |
- |
- |
| 18 |
Hb_100821_010 |
0.3754282965 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 19 |
Hb_002928_080 |
0.3772344629 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g42310, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002392_060 |
0.3774584557 |
- |
- |
gag/pol protein [Bryonia dioica] |