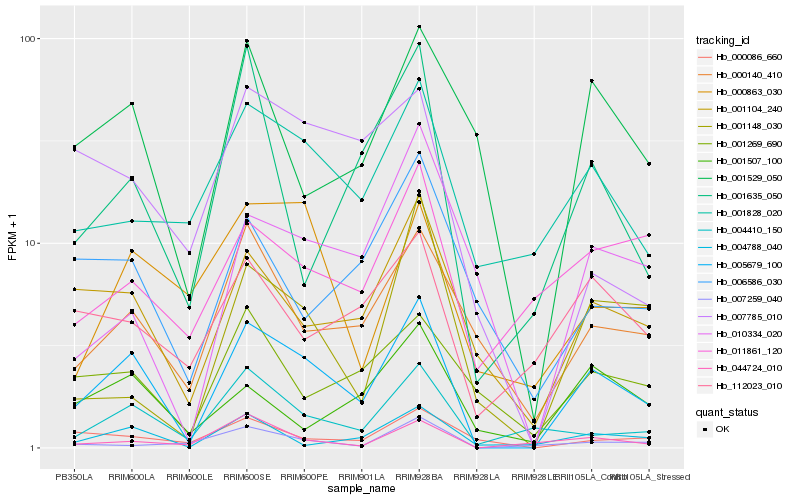
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005679_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s15630g [Populus trichocarpa] |
| 2 |
Hb_001104_240 |
0.219236755 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_004410_150 |
0.2301394526 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 4 |
Hb_004788_040 |
0.2366416504 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000140_410 |
0.2390770871 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 6 |
Hb_044724_010 |
0.2492239066 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 7 |
Hb_001529_050 |
0.252313859 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 8 |
Hb_001828_020 |
0.2538841133 |
- |
- |
hypothetical protein CISIN_1g010132mg [Citrus sinensis] |
| 9 |
Hb_001635_050 |
0.2558517472 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Nicotiana sylvestris] |
| 10 |
Hb_001269_690 |
0.2588334372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_112023_010 |
0.2609149683 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 12 |
Hb_000863_030 |
0.261800158 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 13 |
Hb_000086_660 |
0.261822058 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 14 |
Hb_001148_030 |
0.2637179853 |
- |
- |
- |
| 15 |
Hb_001507_100 |
0.2684303391 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_010334_020 |
0.2702800783 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Sesamum indicum] |
| 17 |
Hb_011861_120 |
0.2715046454 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 18 |
Hb_007259_040 |
0.2719868288 |
- |
- |
hypothetical protein VITISV_005492 [Vitis vinifera] |
| 19 |
Hb_006586_030 |
0.2720049679 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 20 |
Hb_007785_010 |
0.2731576027 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |