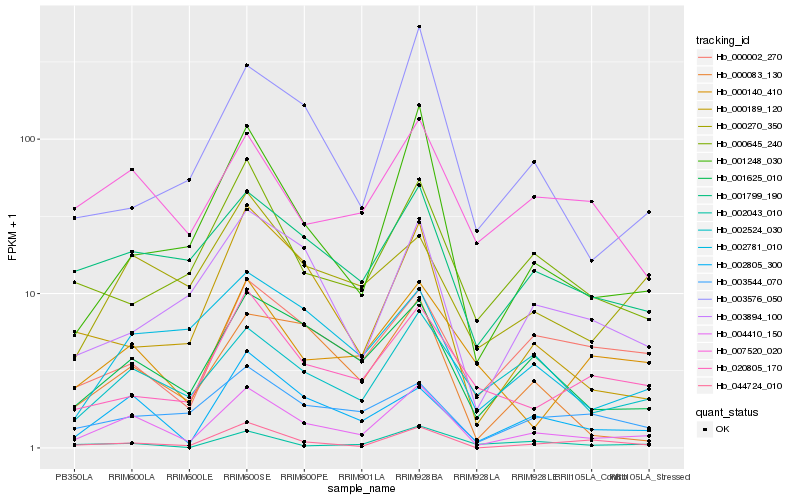
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004410_150 |
0.0 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 2 |
Hb_001625_010 |
0.1605865352 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 3 |
Hb_002805_300 |
0.1625903115 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001799_190 |
0.1792564852 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 5 |
Hb_001248_030 |
0.1822685442 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_044724_010 |
0.1825310181 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 7 |
Hb_002524_030 |
0.1887183687 |
- |
- |
- |
| 8 |
Hb_000002_270 |
0.19663194 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 9 |
Hb_000083_130 |
0.200053131 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 10 |
Hb_007520_020 |
0.2017286125 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
1-deoxy-D-xylulose-5-phosphate reductoisomerase [Hevea brasiliensis] |
| 11 |
Hb_000270_350 |
0.2048187387 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 12 |
Hb_020805_170 |
0.2050676638 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 13 |
Hb_002043_010 |
0.2058242201 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000645_240 |
0.2073476185 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 15 |
Hb_000189_120 |
0.2074190336 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 16 |
Hb_003894_100 |
0.2092801235 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 17 |
Hb_002781_010 |
0.2093611748 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000140_410 |
0.2093975001 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003544_070 |
0.2103558099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003576_050 |
0.2106999101 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |