| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_114146_010 |
0.2019501742 |
- |
- |
hypothetical protein JCGZ_15608 [Jatropha curcas] |
| 3 |
Hb_005305_190 |
0.2340793955 |
transcription factor |
TF Family: NAC |
- |
| 4 |
Hb_006535_010 |
0.2433372757 |
- |
- |
- |
| 5 |
Hb_008221_120 |
0.2609884057 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g63170-like [Jatropha curcas] |
| 6 |
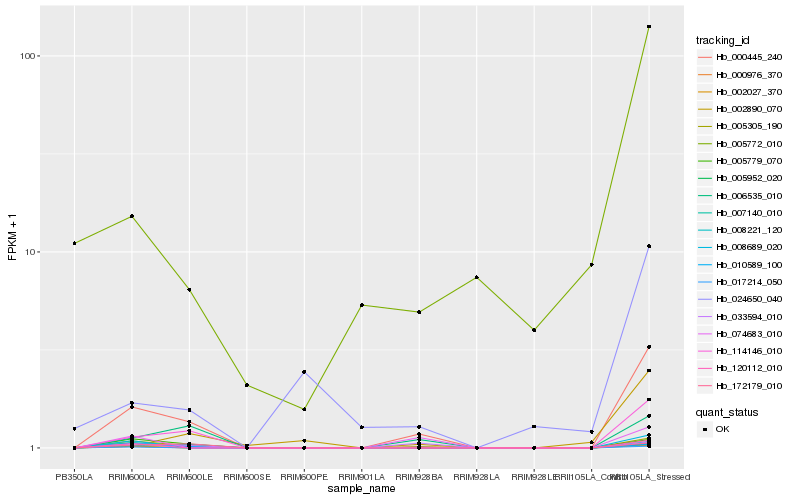
Hb_005779_070 |
0.3088840503 |
- |
- |
- |
| 7 |
Hb_005952_020 |
0.3106391653 |
- |
- |
- |
| 8 |
Hb_008689_020 |
0.3129982646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007140_010 |
0.3137920118 |
- |
- |
- |
| 10 |
Hb_172179_010 |
0.3138657692 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 11 |
Hb_120112_010 |
0.3142707016 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_000976_370 |
0.3158694072 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 13 |
Hb_017214_050 |
0.3209758225 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_002027_370 |
0.3215522685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_033594_010 |
0.3238053144 |
- |
- |
- |
| 16 |
Hb_010589_100 |
0.3279693418 |
- |
- |
Thioredoxin domain-containing protein 9-like protein [Morus notabilis] |
| 17 |
Hb_074683_010 |
0.3336447883 |
- |
- |
- |
| 18 |
Hb_002890_070 |
0.3517636792 |
- |
- |
ATP-dependent RNA helicase family protein [Populus trichocarpa] |
| 19 |
Hb_005772_010 |
0.3525796956 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 20 |
Hb_024650_040 |
0.3650958173 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |