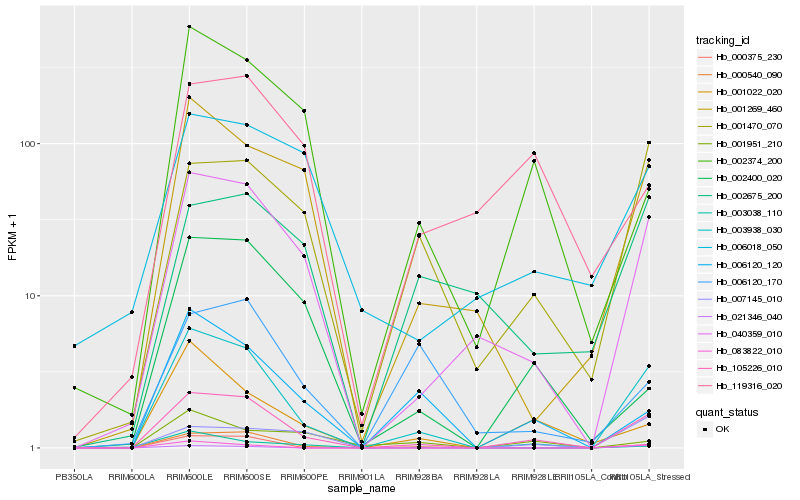
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105226_010 |
0.0 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 2 |
Hb_003938_030 |
0.1511018483 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 3 |
Hb_040359_010 |
0.158027757 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 4 |
Hb_000540_090 |
0.2185198727 |
- |
- |
PREDICTED: chitinase 2-like [Jatropha curcas] |
| 5 |
Hb_001269_460 |
0.2370817862 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 6 |
Hb_001470_070 |
0.2443981238 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_002374_200 |
0.2520326355 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 8 |
Hb_006120_120 |
0.2523474569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000375_230 |
0.2553596611 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 10 |
Hb_001022_020 |
0.2563179433 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 11 |
Hb_007145_010 |
0.2613689398 |
- |
- |
ammonium transporter 3.1-like protein [Camellia sinensis] |
| 12 |
Hb_002400_020 |
0.2656243627 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 13 |
Hb_006018_050 |
0.2664076228 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 14 |
Hb_083822_010 |
0.266968153 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_119316_020 |
0.2821268599 |
- |
- |
- |
| 16 |
Hb_002675_200 |
0.2889146701 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 17 |
Hb_001951_210 |
0.2901548725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_021346_040 |
0.2974989957 |
- |
- |
hypothetical protein JCGZ_26751 [Jatropha curcas] |
| 19 |
Hb_003038_110 |
0.300812056 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 20 |
Hb_006120_170 |
0.3008275585 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108-like [Populus euphratica] |