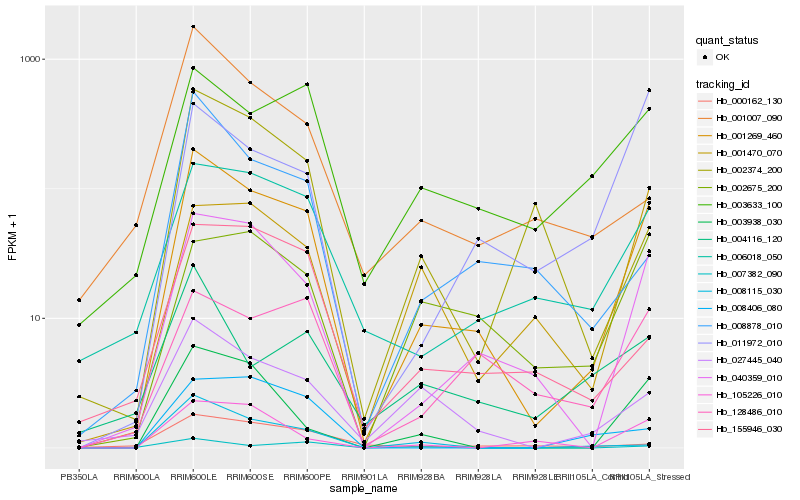
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_460 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 2 |
Hb_040359_010 |
0.1543367352 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 3 |
Hb_006018_050 |
0.2042854447 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 4 |
Hb_003938_030 |
0.2124018033 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 5 |
Hb_027445_040 |
0.2145364446 |
- |
- |
PREDICTED: uncharacterized protein LOC105117996 [Populus euphratica] |
| 6 |
Hb_003633_100 |
0.221497207 |
- |
- |
hypoia-responsive family protein 1 [Hevea brasiliensis] |
| 7 |
Hb_105226_010 |
0.2370817862 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 8 |
Hb_128486_010 |
0.2398175985 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 9 |
Hb_008878_010 |
0.242731655 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 10 |
Hb_155946_030 |
0.2477137648 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 11 |
Hb_002675_200 |
0.2555299422 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 12 |
Hb_007382_090 |
0.2576065529 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_002374_200 |
0.2582188182 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 14 |
Hb_011972_010 |
0.2583894676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008115_030 |
0.2587310567 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 16 |
Hb_008406_080 |
0.2625425333 |
- |
- |
Nonspecific lipid-transfer protein 3 precursor, putative [Ricinus communis] |
| 17 |
Hb_001007_090 |
0.2632670496 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 18 |
Hb_000162_130 |
0.2638014042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001470_070 |
0.2641283504 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_004116_120 |
0.2684458521 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |