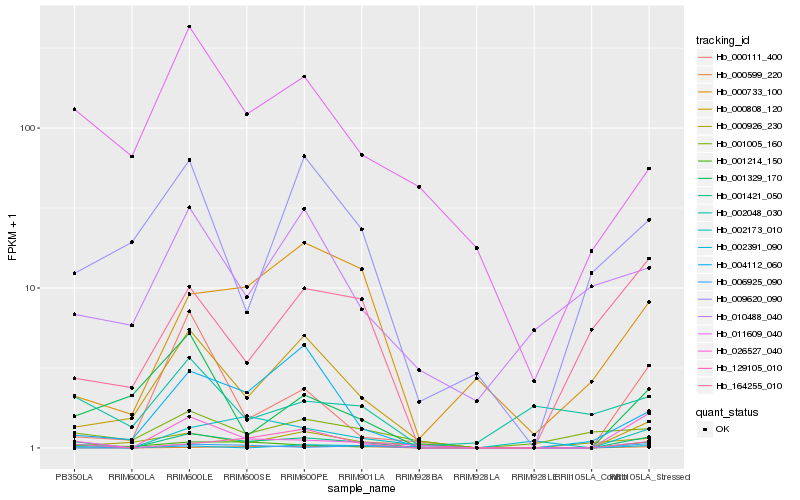
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001421_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_026527_040 |
0.259454149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_129105_010 |
0.2999935417 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_004112_060 |
0.3130932094 |
- |
- |
hypothetical protein POPTR_0017s01950g, partial [Populus trichocarpa] |
| 5 |
Hb_002173_010 |
0.3139251581 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 6 |
Hb_164255_010 |
0.3163890471 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 7 |
Hb_001005_160 |
0.331143115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011609_040 |
0.3312136691 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 9 |
Hb_000733_100 |
0.3342057668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006925_090 |
0.3354366017 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 11 |
Hb_000808_120 |
0.3359378527 |
- |
- |
- |
| 12 |
Hb_000111_400 |
0.343009044 |
- |
- |
hypothetical protein POPTR_0003s19310g [Populus trichocarpa] |
| 13 |
Hb_009620_090 |
0.3477095142 |
- |
- |
hypothetical protein L484_000980 [Morus notabilis] |
| 14 |
Hb_002391_090 |
0.3497088683 |
- |
- |
PREDICTED: IST1-like protein [Jatropha curcas] |
| 15 |
Hb_000599_220 |
0.3525378658 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 16 |
Hb_001329_170 |
0.3529903209 |
- |
- |
Chlorophyll a-b binding protein, chloroplastic [Aegilops tauschii] |
| 17 |
Hb_010488_040 |
0.357476436 |
- |
- |
PREDICTED: phosphomannomutase [Populus euphratica] |
| 18 |
Hb_002048_030 |
0.3614938732 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000926_230 |
0.361894471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001214_150 |
0.3634239399 |
- |
- |
PREDICTED: uncharacterized protein LOC102624288 [Citrus sinensis] |