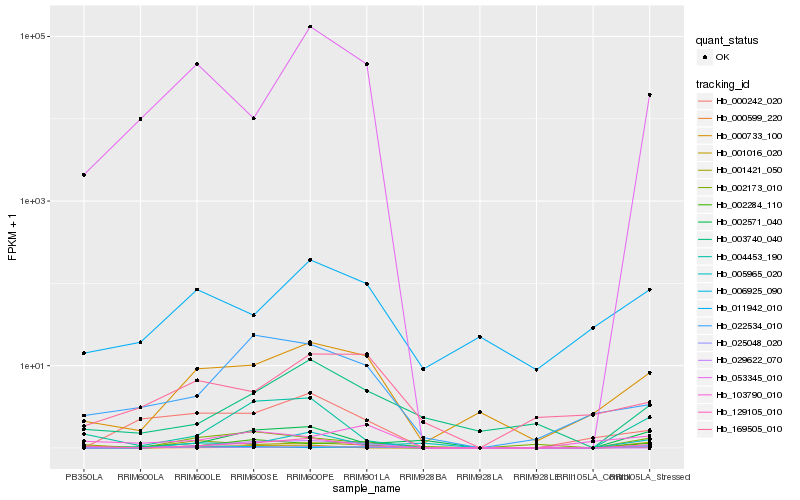
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 2 |
Hb_002284_110 |
0.1252405735 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP18, chloroplastic isoform X2 [Solanum lycopersicum] |
| 3 |
Hb_000733_100 |
0.2546559685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_129105_010 |
0.2837527022 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002173_010 |
0.3157891495 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 6 |
Hb_002571_040 |
0.3217060926 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 7 |
Hb_006925_090 |
0.3271087353 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 8 |
Hb_025048_020 |
0.3371687272 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_022534_010 |
0.340163949 |
- |
- |
hypothetical protein CICLE_v10000066mg [Citrus clementina] |
| 10 |
Hb_004453_190 |
0.3420433367 |
- |
- |
- |
| 11 |
Hb_029622_070 |
0.3426451515 |
- |
- |
- |
| 12 |
Hb_103790_010 |
0.3438728185 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 13 |
Hb_001421_050 |
0.3525378658 |
- |
- |
- |
| 14 |
Hb_005965_020 |
0.360567551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011942_010 |
0.361879547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001016_020 |
0.3646175133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003740_040 |
0.3664863823 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase isozyme L3, putative [Ricinus communis] |
| 18 |
Hb_000242_020 |
0.3716087591 |
- |
- |
hypothetical protein CICLE_v100246891mg, partial [Citrus clementina] |
| 19 |
Hb_169505_010 |
0.3743636707 |
- |
- |
hypothetical protein JCGZ_04480 [Jatropha curcas] |
| 20 |
Hb_053345_010 |
0.3794768584 |
- |
- |
- |